Oral Session
Data Processing
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| 09:15 | 0425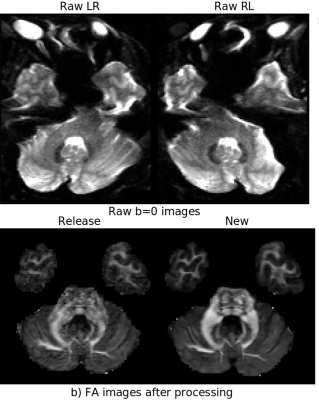 |
ReImagining the Young Adult Human Connectome Project (HCP) Diffusion MRI Dataset
M. Okan Irfanoglu1, Ahmad Beyh2,3, Marco Catani2, Flavio Dell'Acqua2, and Carlo Pierpaoli1
1QMI, NIBIB/NIH, Bethesda, MD, United States, 2Natbrainlab, King's College London, London, United Kingdom, 3Laboratory of Neurobiology, Department of Cell and Developmental Biology, University College London, London, United Kingdom
The Human Connectome Project (HCP) has brought significant advancements in hardware, acquisition, and preprocessing. Even after a decade since its collection, the HCP diffusion MRI data is still relevant for its richness and high resolution. Noise and geometric distortions, however, are particularly pronounced in this dataset. In this work, we have reprocessed nearly the entire HCP dMRI dataset while applying several recent processing improvements. We compared the quality of the newly processed dMRI outputs to the release version. We observed clearly detectable improvements. The data originated from this new processing will be made publicly available.
|
|
| 09:27 | 0426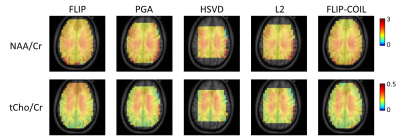 |
A coil-based approach to accelerate the FLIP spatial lipid removal algorithm in 3D EPSI. Video Permission Withheld
Peter Adany1, In-Young Choi1,2,3,4, and Phil Lee1,3,4
1Hoglund Biomedical Imaging Center, University of Kansas Medical Center, Kansas City, KS, United States, 2Department of Neurology, University of Kansas Medical Center, Kansas City, KS, United States, 3Department of Radiology, University of Kansas Medical Center, Kansas City, KS, United States, 4Department of Molecular & Integrative Physiology, University of Kansas Medical Center, Kansas City, KS, United States
We have recently developed the spatial domain, Fast LIpid signal Processing (FLIP) algorithm to remove subcutaneous lipid signals in MRSI. Practical application of FLIP to 3D EPSI was challenging due to the need for long processing times. We present an updated algorithm, named FLIP-COIL, which significantly reduces the processing times utilizing receive coil senstivity profiles. Algorithms including FLIP, FLIP-COIL, as well as PGA, HSVD and L2 are compared in 3D EPSI of ten subjects. This study demostrates that the FLIP-COIL approach can drastically reduce processing time with favorable performance of lipid removal in 3D EPSI over existing other algorithms.
|
|
| 09:39 | 0427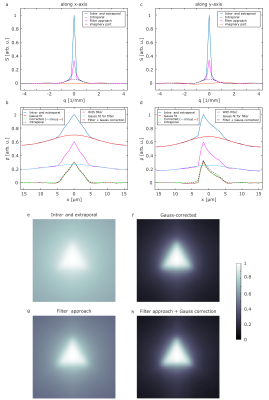 |
Correction of diffusion pore imaging (DPI) data in the presence of extraporal water and transmembrane water exchange
Dominik Ludwig1,2, Frederik B. Laun3, Karel D. Klika4, Julian Rauch1,2, Mark E. Ladd1,2,5, Peter Bachert1,2, and Tristan A. Kuder1
1Deparment of Medical Physics in Radiology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Faculty of Physics and Astronomy, Heidelberg University, Heidelberg, Germany, 3Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 4Molecular Structure Analysis, German Cancer Research Center (DKFZ), Heidelberg, Germany, 5Faculty of Medicine, Heidelberg University, Heidelberg, Germany
Diffusion pore imaging might open a new window into pathologies by providing histology-like information non-invasively by estimating pore size and shape distributions. For current pore imaging approaches, closed pores filled with an NMR visible diffusing medium are assumed, while extraporal components and exchange are neglected, which limits applicability. We propose a method based on Gaussian phase approximation to suppress effects of extraporal fluids and transmembrane water exchange and compare the approach to a filter-based method. Thus, one main obstacle for in vivo applications is reduced, the required high gradient amplitudes may be obtained using local gradient coils in the future.
|
|
| 09:51 | 0428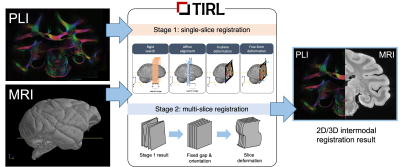 |
Deformable Registration of Serial Micrographs to Post-Mortem MRI in the BigMac Dataset Using TIRL
Istvan N Huszar1, Adele Smart1, Saad Jbabdi1, Mark Jenkinson1, Amy FD Howard1, and Karla L Miller1
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom The BigMac dataset combines extensive post-mortem MRI with histology and polarised light imaging (PLI) in a macaque brain. Adding a new extension to our recently developed platform, the Tensor Image Registration Library (TIRL), we address unique challenges associated with this dataset and successfully align 77 2-D polarised light micrographs to high-resolution structural MRI data. |
|
| 10:03 | 0429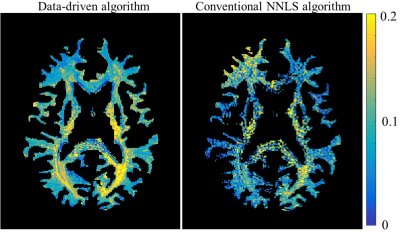 |
Improving myelin water mapping using a new data-driven paradigm for multicomponent analysis of T2 relaxation times
Noam Omer1, Neta Stern1, Tamar Blumenfeld-Katzir1, Chen Solomon1, Meirav Galun2, and Noam Ben-Eliezer1,3,4
1Biomedical Engineering, Tel Aviv University, Tel Aviv, Israel, 2Department of Computer Science and Applied Mathematics, Weizmann Institute of Science, Rehovot, Israel, 3Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 4Center for Advanced Imaging Innovation and Research (CAI2R), New-York University Langone Medical Center, New York, NY, United States
The common approach to myelin mapping relies on multi-T2 component (mcT2) analysis, where the signal from a single voxel is separated into its underlying distribution of T2 values. This approach is highly challenging due to the ill posedness of extracting multiple free parameters from a single voxel data. We present a new data-driven paradigm, where the white matter is first analyzed to identify a finite set of multi-T2 distributions, which are then used to locally analyze the signal in each voxel. Application on white matter tissue produced improved myelin quantifications, without a priory fixing the number of sub-voxel compartments.
|
|
10:15 |
0430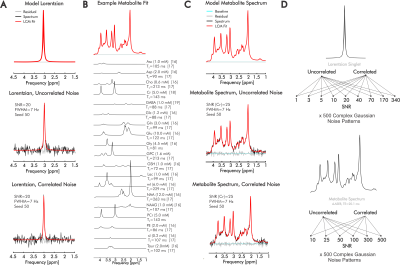 |
Complex fitting of 1H-MR spectra improves quantification precision independent of SNR and noise correlation
Leonardo Campos1, Kelley M. Swanberg1, Martin Gajdošík1, Karl Landheer1, and Christoph Juchem1,2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Radiology, Columbia University, New York, NY, United States
Quantification of in vivo proton magnetic resonance spectra (1H-MRS) still commonly involves evaluation of exclusively the real part of acquired spectral signals, but ignoring the information contained in the imaginary component may limit precise identification of individual metabolite contributions. Here, we assess quantification precision relative to SNR and noise correlation for both real and complex linear combination model fits of simulated 1H-MRS spectra reflecting brain metabolite concentrations and T2. Extending our results to inclusion of measured in vivo baselines, we demonstrate consistent improvements in metabolite quantification precision and/or accuracy by complex relative to real fits.
|
|
| 10:27 | 0431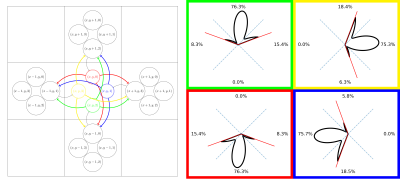 |
Connectivity matrix from a fODF weighted graph: An alternative to probabilistic tractography
Michael Paquette1, Cornelius Eichner1, and Alfred Anwander1
1Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany
We introduce a computationally efficient fODF-weighted graph structure where shortest-paths through white matter compute the probability of connection while naturally limiting the angle of propagation between steps. Connectivity matrices obtained from this structure maintain many properties of probabilistic streamline count connectomes while avoiding the sampling bias of tractography.
|
|
| 10:39 | 0432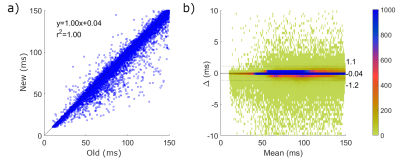 |
Fast inline reconstruction of T2 maps from dual echo spin echo images
Jeff Snyder1, Peter Seres1, Robert W Stobbe1, Justin G Grenier1, Penelope Smyth2, Gregg Blevins2, and Alan H Wilman1
1Biomedical Engineering, University of Alberta, Edmonton, AB, Canada, 2Department of Medicine, Division of Neurology, University of Alberta, Edmonton, AB, Canada
A fast reconstruction method for PD-T2w T2 maps is presented and compared to the previous L2 norm minimization technique using Bland-Altman analysis in a five patient multiple sclerosis data set acquired at 3 T. The new (subtraction) technique was in excellent agreement with the L2 norm method (confidence intervals of -1.1 to +1.2 ms), with average single slice reconstruction times of 0.6 s compared to 134 s. The speed of T2 map production allowed accurate (based on sequence simulation via Bloch equations) inline T2 maps directly on the MRI console.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.