Oral Session
Deep/Machine Learning-Based Image Analysis
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| 16:45 | 0551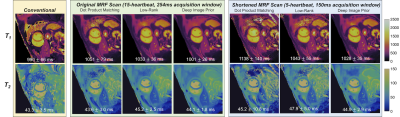 |
Accelerated 2D Cardiac MRF Using a Self-Supervised Deep Image Prior Reconstruction
Jesse Ian Hamilton1,2
1Radiology, University of Michigan, Ann Arbor, MI, United States, 2Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
A physics-based deep learning reconstruction is proposed for cardiac MRF that is based on the deep image prior framework, which employs self-supervised training and does not require additional in vivo training data. Calculation of the forward model is accelerated using a neural network to rapidly output MRF signal timecourses (in place of a Bloch simulation) and low-rank modeling to reduce the number of NUFFT operations. The proposed reconstruction enables cardiac T1/T2 mapping during a shortened 5-heartbeat breathhold and 150ms acquisition window.
|
|
16:57 |
0552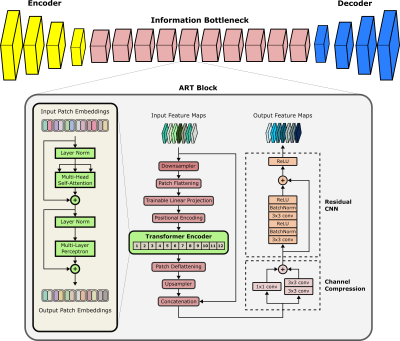 |
Cycle-Consistent Adversarial Transformers for Unpaired MR Image Translation
Onat Dalmaz1,2, Mahmut Yurt3, Salman UH Dar1,2, and Tolga Çukur1,2,4
1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center (UMRAM), Bilkent University, Ankara, Turkey, 3Electrical Engineering, Stanford University, Stanford, CA, United States, 4Neuroscience Program, Aysel Sabuncu Brain Research Center, Bilkent University, Ankara, Turkey Translating acquired sequences to missing ones in multi-contrast MRI protocols can dramatically reduce scan costs. Neural network models devised for this purpose are characteristically trained on paired datasets, which can be difficult to compile. Moreover, these models exclusively rely on convolutional operators with undesirable biases towards feature locality and spatial invariance. Here, we present a cycle-consistent translation model, ResViT, to enable training on unpaired datasets. ResViT combines localization power of convolution operators with contextual sensitivity of transformers. Demonstrations on multi-contrast MRI datasets indicate the superiority of ResViT against state-of-the-art translation models. |
|
17:09 |
0553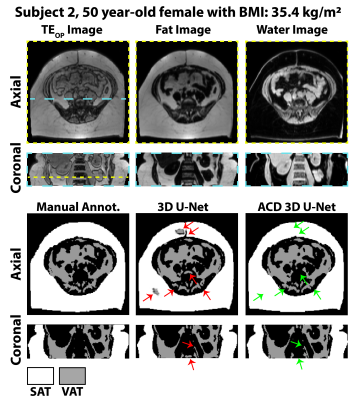 |
Automated Adipose Tissue Segmentation using 3D Attention-Based Competitive Dense Networks and Volumetric Multi-Contrast MRI
Sevgi Gokce Kafali1,2, Shu-Fu Shih1,2, Xinzhou Li1, Shilpy Chowdhury3, Spencer Loong4, Samuel Barnes3, Zhaoping Li5, and Holden H. Wu1,2
1Radiological Sciences, University of California, Los Angeles, Los Angeles, CA, United States, 2Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States, 3Radiology, Loma Linda University Medical Center, Loma Linda, CA, United States, 4Psychology, Loma Linda University School of Mental Health, Loma Linda, CA, United States, 5Medicine, University of California, Los Angeles, Los Angeles, CA, United States
Subcutaneous and visceral adipose tissue (SAT/VAT) are potential biomarkers to detect future risks of metabolic diseases. However, the current standard for analysis relies on manual annotations that require expert knowledge and are time-consuming. Previous neural networks for automatically segmenting adipose tissue had suboptimal performance for VAT. This work developed a new 3D attention-based competitive dense network to rapidly (84 ms/slice) and accurately segment SAT/VAT in adults with obesity by leveraging multi-contrast MRI inputs and considering the complex VAT features. The new network achieved high Dice scores (>0.96) and accurate volume measurements (difference<1.6%) for SAT/VAT with respect to manual annotations.
|
|
| 17:21 | 0554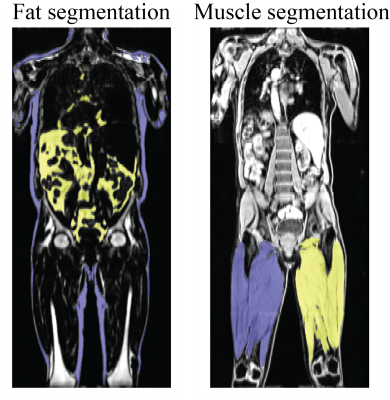 |
An End-to-End Segmentation Pipeline for Dixon Adipose and Muscle by Neural Nets (DAMNN) Video Permission Withheld
Karl Landheer1, Jonathan Marchini1, Benjamin Geraghty1, Prodromos Parasoglou1, Stefanie Hectors1, Farshid Sepehrband1, Nicholas Gale1, Andrew Murphy1, Johnathon Walls1, and Mary Germino1
1Regeneron Pharmaceuticals, Inc, Tarrytown, NY, United States
An end-to-end pipeline was developed that processes whole-body Dixon MRI data sets from UK Biobank and corrects for overlapping slices, inhomogeneous signal intensities, and fat-water swaps to produce high quality 3D data sets. Segmentation maps for subcutaneous/visceral fat and left/right thigh muscles from these 3D data sets were then produced using neural networks, and muscle and fat volume phenotypes were extracted. The Jaccard index for the validation data sets was 93.3% for the fat segmentation and 96.9% for the muscle segmentation. Excellent correspondence was obtained with the extracted muscle and fat volumes and similar metrics from a commercially available package.
|
|
| 17:33 | 0555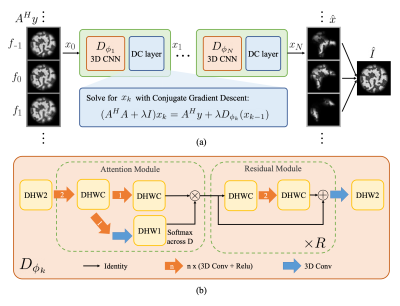 |
ResoNet: Physics Informed Deep Learning based Off-Resonance Correction Trained on Synthetic Data
Alfredo De Goyeneche 1, Shreya Ramachandran1, Ke Wang1,2, Ekin Karasan1, Stella Yu1,2, and Michael Lustig1
1Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 2International Computer Science Institute, University of California, Berkeley, Berkeley, CA, United States
We propose a physics-inspired, unrolled-deep-learning framework for off-resonance correction. Our forward model includes coil sensitivities, multi-frequency bins, and non-uniform Fourier transforms hence compatible with fat/water imaging and parallel imaging acceleration. The network, which includes data-consistency terms and CNN modules serving as proximal operators, is trained end-to-end using only synthetic random field maps, coil sensitivities, and noise-like images with statistics (smoothness) mimicking natural signals. Our aim is to train the network to reverse off-resonance irrespective of the type of imaging, and hence generalizable to any anatomy and contrast without retraining. We demonstrate initial results in simulations, phantom, and in-vivo data.
|
|
| 17:45 | 0556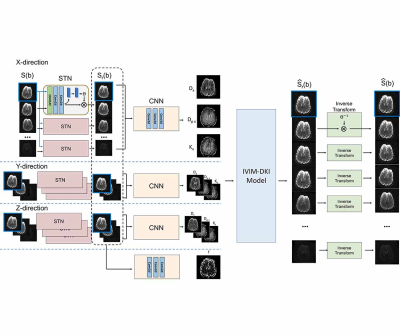 |
Registration and quantification net (RQnet) for IVIM-DKI analysis
Wonil Lee1, Giyong Choi1, Jongyeon Lee1, and HyunWook Park1
1Electrical Engineering, KAIST, Daejeon, Korea, Republic of
Accurate alignment of multiple diffusion-weighted images must be preceded to predict accurate diffusion parameters. A number of registration approaches have been studied (1,2). However, most of them minimize the dissimilarity between diffusion weighted image and a reference, which can cause errors because the characteristics of the images are different. In order to accurately investigate diffusion, perfusion, and kurtosis parameters using hybrid IVIM-DKI model, a deep learning network is proposed as an end-to-end fashion. This method is entirely unsupervised learning, which does not require reference image for registration and the labeled IVIM-DKI parameters for registration and quantification.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.