Power Pitch
Pitch: Image Reconstruction & Signal Models
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

Power Pitch Session: How it Works
1st Hour: 2-minute Power Pitches in the Power Pitch Theater.
2nd Hour: 60-minute digital poster presentations at the smaller screens around the perimeter of the Power Pitch Theater.
09:15 |
0240. Video Not Availablel"> 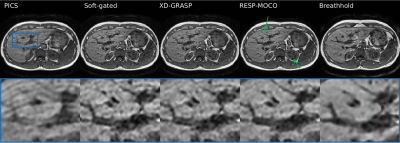 |
Computationally tractable high resolution non-rigid motion compensated 3D abdominal image reconstruction
Tom Bruijnen1,2, Tim Schakel1, and Cornelis AT van den Berg1,2
1Department of Radiotherapy, University Medical Center Utrecht, Utrecht, Netherlands, 2Computational Imaging Group for MR Therapy and Diagnostics, University Medical Center Utrecht, Utrecht, Netherlands
Respiratory and bulk organ motion remains a significant problem in abdominal 3D MRI. Motion compensated image reconstruction is a potential generic solution, but requires solving a large computational problem. In this work we propose to split the image reconstruction into sagittal planes to correct for the majority of respiratory and bulk organ motion. We demonstrate that the proposed method reconstructs high quality free-breathing T1 gradient echo images that are qualitatively comparable to breathhold scans. Future work will assess the feasibility of the method for dynamic contrast enhanced MRI and MR cholangiopancreatography.
|
|
| 09:17 | 0241.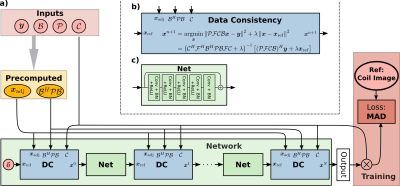 |
Deep Subspace Learning for Improved T1 Mapping using Single-shot Inversion-Recovery Radial FLASH
Moritz Blumenthal1, Xiaoqing Wang1,2, and Martin Uecker1,2,3
1Institute for Diagnostic and Interventional Radiology, University Medical Center Göttingen, Göttingen, Germany, 2DZHK (German Centre for Cardiovascular Research), Göttingen, Germany, 3Institute of Medical Engineering, Graz University of Technology, Graz, Austria
Subspace based reconstructions can be used for quantitative imaging. We extend the recently added neural network framework in BART to support non-Cartesian trajectories and linear subspace constraint reconstructions. The network is trained to reconstruct coefficient maps from single-shot radial FLASH inversion recovery acquisitions. T1 maps are estimated using pixel-wise fitting to the signal model. The reconstruction quality of the coefficient and T1 maps are improved compared to an $$$\ell_1$$$-Wavelet regularized reconstruction. The subspace constraint network can be used for any linear subspace constraint reconstruction.
|
|
09:19 |
0242.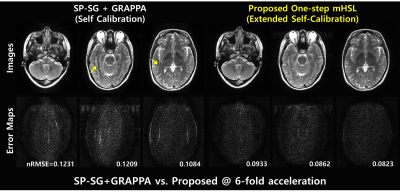 |
Self-Calibrating Aliasing-Controlled Simultaneous Multi-Slice MR Image Reconstruction from Generalized 3D Fourier Encoding Perspective
Eun Ji Lim1, Chul-Ho Sohn2, Taehoon Shin3, and Jaeseok Park1,4
1Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of, 3Division of Mechanical and Biomedical Engineering, Ewha Womans University, Seoul, Korea, Republic of, 4Department of Intelligent Precision Healthcare Convergence, Sungkyunkwan University, Suwon, Korea, Republic of
To jointly resolve inter-slice leakages and in-plane aliasing in undersampled SMS imaging, we proposed a novel, one-step solution for SMS-reconstruction optimally exploiting variable-density(VD) sampling from generalized 3D Fourier encoding perspective. Extended controlled aliasing is performed by upsampling VD sampled data in the kz-direction. A slice-specific null space operator is learned using extended self-calibration including CAIPI slices and in-plane images. Aliasing artifacts are jointly resolved in ky-and kz-directions by balancing null space consistency with a low rank prior while enforcing slice-specific VD-sampled data in 3D k-space. We demonstrated the proposed method outperforms competing methods at SMS=3,R=2 in head and knee.
|
|
| 09:21 | 0243.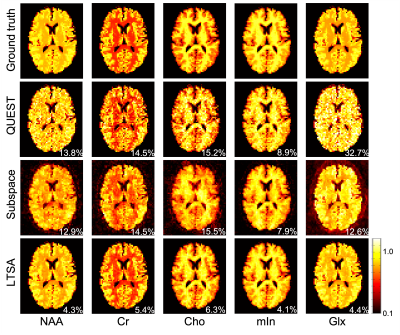 |
MRSI spectral quantification using linear tangent space alignment (LTSA)-based manifold learning
Chao Ma1,2 and Georges El Fakhri1,2
1Radiology, Massachusetts General Hospital, Boston, MA, United States, 2Radiology, Harvard Medical School, Boston, MA, United States
Spectral quantification is a critical step in quantitative MRS/MRSI. The recently proposed subspace-based spectral quantification method represents the spectral distribution of each model as a subspace model and enables effective use of spatiospectral priors to improve parametric estimation. However, modeling spectral distribution of each metabolite as a separate subspace leads to a large number of unknowns, which renders the resultant parametric estimation problem challenging when SNR is low. To address this issue, we propose a new linear tangent space alignment-based method for MRSI quantification, leveraging the intrinsic low-dimensional structure information of the underlying MRSI signals for improved parametric estimation.
|
|
| 09:23 | 0244.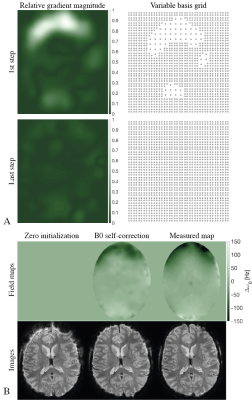 |
Off-Resonance Self-Correction by Implicit Temporal Encoding: Performance at 7T
Franz Patzig1 and Klaas Paul Pruessmann1
1Institut for Biomedical Engineering, ETH Zurich and University of Zurich, Zurich, Switzerland
B0-induced distortions are a main caveat for trajectories with long readout durations, particularly at high field. Recently, B0 self-correction based on implicit temporal encoding by the coil array has been proposed and shown to be feasible at 3T. This work focuses on application of the approach at 7T. Performance analysis show that due to the increased strength of B0 inhomogeneities constant density bases to model B0 might not be sufficient. A variable density basis adapting to the gradient is proposed yielding spatially-dependent regularization. By means of this, reconstruction results can be improved demonstrating the feasibility of B0 self-correction at 7T.
|
|
| 09:25 | 0245.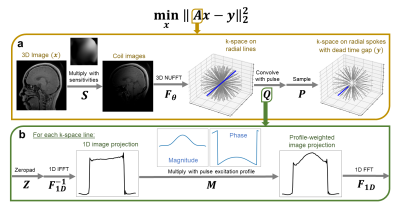 |
T1-weighted ZTE MRI with phase-modulated RF and a parallel imaging, inverse problem-based reconstruction
Shreya Ramachandran1 and Michael Lustig1
1Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States
For fast T1-weighted imaging, zero echo time (ZTE) imaging provides rapid sampling of 3D k-space, but lacks T1 contrast due to its small flip angle excitation with short hard pulses. To efficiently increase T1 contrast, longer phase-modulated pulses can be used. However, longer pulses lead to larger dead-time gaps and pulse profile-weighted images. Here, we formulate an inverse problem to directly reconstruct profile-compensated images from multi-coil data without any intermediate step for dead-time infilling. We demonstrate that leveraging coil sensitivities and alternating phase-modulated excitations sufficiently condition the inverse problem, allowing for iterative reconstructions of T1-weighted acquisitions.
|
|
| 09:27 | 0246.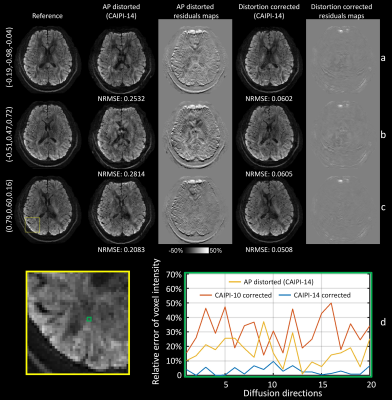 |
Integration of blip reversal with CAIPI sampling enables simultaneous correction of slice aliasing and distortion in 3D multi-slab diffusion MRI
Ziyu Li1, Karla L. Miller1, and Wenchuan Wu1
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
3D EPI can provide optimal SNR efficiency for high-resolution diffusion MRI but is prone to aliasing of edge slices and in-plane distortion artifacts. We propose a highly-efficient method for correcting slice-aliasing and distortion for 3D multi-slab imaging without increasing scan time. Blip reversal is integrated into CAIPI sampling within a single scan. In addition, we extend the FOV along the slice direction to remove slice-aliasing artifacts. Field maps are estimated from the blip-reversed data and incorporated in the final joint reconstruction. The efficacy of the method is quantitatively and systematically validated using highly realistic simulations.
|
|
09:29 |
0247.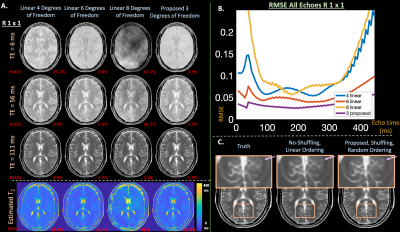 |
Learning compact latent representations of signal evolution for improved shuffling reconstruction
Yamin Arefeen1, Junshen Xu1, Molin Zhang1, Jacob White1, Berkin Bilgic2,3, and Elfar Adalsteinsson1,4,5
1Massachusetts Institute of Technology, Cambridge, MA, United States, 2Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 3Department of Radiology, Harvard Medical School, Boston, MA, United States, 4Harvard-MIT Health Sciences and Technology, Cambridge, MA, United States, 5Institute for Medical Engineering and Science, Cambridge, MA, United States
Applying linear subspace constraints in the shuffling forward model enables reconstruction of signal dynamics through reduced degrees of freedom. Other techniques train auto-encoders to learn latent representations of signal evolution to apply as regularization. This work inserts the decoder portion of an auto-encoder directly into the shuffling forward model to reduce degrees of freedom in comparison to linear techniques. We show that auto-encoders represent fast-spin-echo signal evolution with 1 latent variable, in comparison to 3-4 linear coefficients. Then, the reduced degrees of freedom enabled by the decoder improves reconstruction results in comparison to linear constraints in simulation and in-vivo experiments.
|
|
| 09:31 | 0248.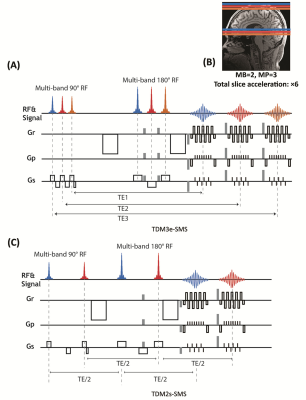 |
Integrating time division multiplexing (TDM) and simultaneous multi-slice (SMS) for accelerating combined diffusion-relaxometry
Yang Ji1,2, William Scott Hoge3, Borjan Gagoski4, Carl-Fredrik Westin 3, Yogesh Rathi1,3, and Lipeng Ning1
1Department of Psychiatry, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States, 2Wellcome Centre for Integrative Neuroimaging, FMRIB Division, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 3Department of Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States, 4Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Harvard Medical School, Boston, MA, United States
We introduce an MRI sequence that integrates the time-division multiplexing (TDM) technique and simultaneous multi-slice (SMS) method to achieve a high slice-acceleration (~6x) factor for acquiring relaxation-diffusion MRI. Two variants of the sequence, i.e., TDM3e-SMS and TDM2s-SMS, were developed to simultaneously acquire slice groups with 3 distinct echo times (TEs) and 2 slice groups with the same TE, respectively. Both sequences were evaluated on a 3T scanner with in-vivo human brains and compared with standard single-band (SB) EPI and SMS-EPI using diffusion measures and tractography results. Results have shown that TDM-SMS provides reliable measures for relaxation-diffusion and standard diffusion MRI.
|
|
| 09:33 | 0249.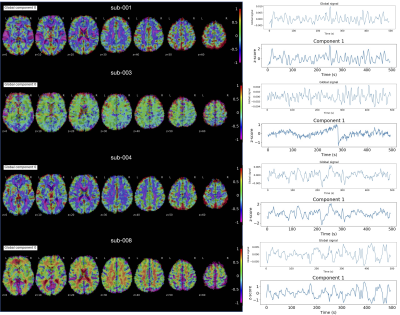 |
A multi-echo low-rank and sparse algorithm that reduces the bias of global fluctuations on the estimation of neuronal signal
Eneko Uruñuela1, Stefano Moia1, and César Caballero-Gaudes1
1Basque Center on Cognition, Brain and Language, Donostia - San Sebastián, Spain
This work introduces a novel multi-echo fMRI deconvolution approach that reduces the effect of global fluctuations (e.g., motion effects, physiological confounds, artefacts) on blindly mapping the brain’s response to single-trial BOLD events without prior timing information. The new sparse-plus-low-rank multi-echo multivariate paradigm free mapping (SPLORA) algorithm is compared with a trial-based known-timing GLM analysis and its predecessor multivariate multiecho paradigm free mapping (MvME-PFM) approach. This method allows exploring the brain’s functional dynamics during task, naturalistic and resting-state paradigms, being less affected by motion and physiological confounds, thus avoiding global signal regression to estimate neuronal related activity with multi-echo fMRI.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.