Power Pitch
Pitch: MR Fingerprinting
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

Power Pitch Session: How it Works
1st Hour: 2-minute Power Pitches in the Power Pitch Theater.
2nd Hour: 60-minute digital poster presentations at the smaller screens around the perimeter of the Power Pitch Theater.
16:45 |
0557.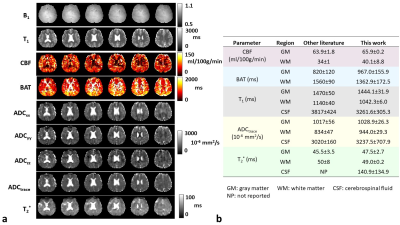 |
Simultaneous perfusion, diffusion, T2*, and T1 mapping with MR Fingerprinting (MRF-PDT)
Hongli Fan1,2, Lisa Bunker3, Alexandra Zezinka Durfee3, Xiaohong Joe Zhou4, Argye E. Hillis3, and Hanzhang Lu1,2,5
1Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2The Russell H. Morgan Department of Radiology & Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 4Center for Magnetic Resonance Research and Department of Radiology, University of Illinois at Chicago, Chicago, IL, United States, 5F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
Quantitative mapping of brain perfusion, diffusion, T2*, and T1 has a broad range of clinical applications. The present work aims to develop a novel pulse sequence that can simultaneously map perfusion, diffusion, T2* and T1 with MR fingerprinting, dubbed MRF-PDT, with a total scan time of <6 minutes. This technique was first demonstrated on healthy volunteers, then on two patients with ischemic stroke. All maps derived from MRF-PDT exhibited the expected image contrasts with quantitative values consistent with those reported in literature. In addition, test-retest studies confirmed the reproducibility of the proposed technique.
|
|
| 16:47 | 0558.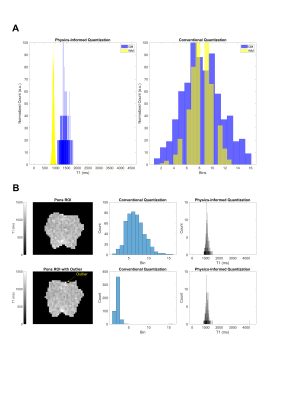 |
Improving Radiomics Reproducibility Using MR Fingerprinting and Physics-Informed Quantization
Walter Zhao1,2, Zheyuan Hu1, Anahita Fathi Kazerooni3,4, Gregor Körzdörfer5, Matthias Nittka5, Christos Davatzikos3,4, Satish E. Viswanath1, Xiaofeng Wang6, Chaitra Badve7, and Dan Ma1
1Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Medical Scientist Training Program, Case Western Reserve University, Cleveland, OH, United States, 3Center for Biomedical Image Computing and Analysis, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 4Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 5Siemens Healthineers, Erlangen, Germany, 6Department of Quantitative Health Sciences, Cleveland Clinic, Cleveland, OH, United States, 7Department of Radiology, Case Western Reserve University and University Hospitals Cleveland Medical Center, Cleveland, OH, United States
MR fingerprinting (MRF) is a rapid, quantitative imaging approach with significant potential for use in clinical studies, including radiomic applications. Due to its quantitative nature, robustness to system imperfections, and requiring fewer image preprocessing steps, we believe MRF radiomics is uniquely positioned to offer improved reproducibility and generalizability compared to conventional MRI. Here we report reproducibility results of MRF T1 and T2 radiomic features in the healthy human brain, and introduce a novel physics-informed quantization approach for improved reproducibility of quantitative image texture features.
|
|
| 16:49 | 0559.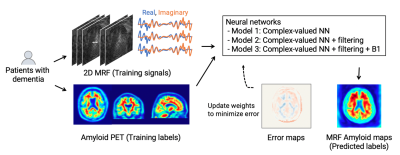 |
MR fingerprinting and complex-valued neural network for quantification of brain amyloid burden
Shohei Fujita1,2, Yujiro Otsuka1,3,4, Katsutoshi Murata5, Gregor Koerzdoerfer6, Mathias Nittka6, Yumiko Motoi7,8, Madoka Nakajima8,9, Koji Murakami10, Issei Fukunaga1, Koji Kamagata1, Osamu Abe2, and Shigeki Aoki1
1Department of Radiology, Juntendo University, Tokyo, Japan, 2Department of Radiology, The University of Tokyo, Tokyo, Japan, 3Milliman Inc, Tokyo, Japan, 4Plusman LLC, Tokyo, Japan, 5Siemens Healthcare Japan KK, Tokyo, Japan, 6Siemens Healthcare GmbH, Erlangen, Germany, 7Department of Neurology, Juntendo University, Tokyo, Japan, 8Medical Center for Dementia, Juntendo University, Tokyo, Japan, 9Department of Neurosurgery, Juntendo University, Tokyo, Japan, 10Division of Nuclear Medicine, Department of Radiology, Juntendo University, Tokyo, Japan
We developed a framework utilizing MR fingerprinting and a complex-valued neural network to detect brain amyloid burden. The tailored neural network was trained on real amyloid-PET imaging data and MR fingerprinting acquisitions to estimate PET-derived amyloid deposition from the MR fingerprinting signal evolutions. This complex-valued neural network architecture, designed to increase sensitivity to amyloid-related signals, showed a subject-level amyloid positivity classification with AUC = 0.87 in patients with cognitive decline. The proposed method enables non-invasive amyloid burden mapping, T1 and T2 mapping in a single scan, and is suitable not only for diagnosis but also for monitoring amyloid-reducing treatments.
|
|
| 16:51 | 0560.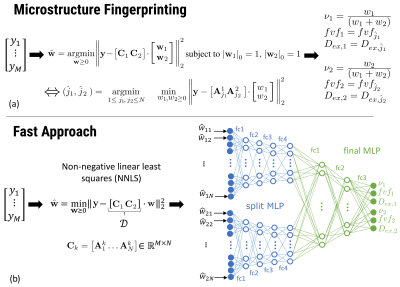 |
Fast multi-compartment microstructure fingerprinting using deep neural networks
Quentin Dessain1, Nicolas Delinte1, Benoit Macq1, and Gaëtan Rensonnet1
1ICTEAM, Université Catholique de Louvain, Louvain La Neuve, Belgium We estimate microstructural features of crossing fascicles in the white matter by using a fast multi-compartment fingerprinting, an extension of MR fingerprinting to diffusion MRI. The acceleration uses efficient sparse optimization and a dedicated feed-forward neural network to circumvent the inherent combinatorial complexity of the fingerprinting estimation. The accuracy of the results and the speedup factors obtained on in vivo brain data suggest the potential of our method for a fast quantitative estimation of microstructural features in complex white matter configurations. |
|
16:53 |
0561.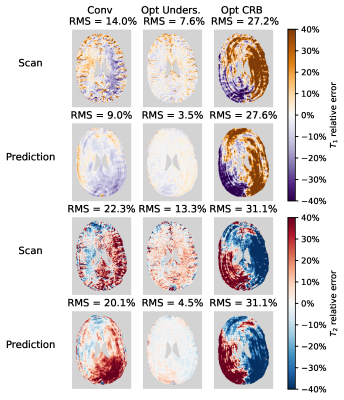 |
Sequence optimisation mitigating undersampling errors in Magnetic Resonance Fingerprinting
David Heesterbeek1, Martin van Gijzen2, Frans Vos1,3, and Martijn Nagtegaal1
1Department of Imaging Physics, Delft University of Technology, Delft, Netherlands, 2Department of Numerical Analysis, Delft University of Technology, Delft, Netherlands, 3Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, Netherlands
In MR Fingerprinting acquisitions, the choice of flip angle sequence has a strong effect on the accuracy of the estimated parameter maps. Undersampling artefacts in time-series images are a dominant source of error for standard MRF estimations. We propose to use an undersampling-error model leveraging on perturbation theory, to optimise flip angle patterns taking into account the k-space readout, a realistic ground truth and signal phase. In vivo scans show strong visible reduction in error. Root-mean-square errors in $$$T_1$$$ and $$$T_2$$$ were reduced with at least 6.4%-points and 9%-points respectively, compared to a Cramér-Rao bound optimised and a conventional pattern.
|
|
| 16:55 | 0562.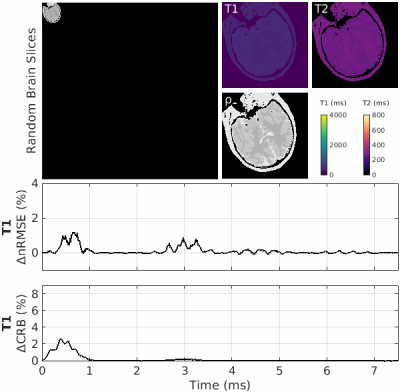 |
Visualizing Encoding Efficiency of MR Fingerprinting Sequences using Leave-One-Out Perturbation (LOOP)
Christian Guenthner1, Johanna Stimm1, and Sebastian Kozerke1
1ETH and University Zurich, Zurich, Switzerland
In Magnetic Resonance Fingerprinting, the accuracy of the results is dominated by undersampling artifacts. While in classical relaxometry techniques, the omission of data always leads to a larger error, in fingerprinting, undersampling artifacts can lead to both an increase or a decrease in error. The “temporal encoding efficiency” of fingerprinting can be analyzed based on the change in matching error upon omission of a single time point (leave-one-out). We propose a first-order perturbation of the undersampling error to visualize and identify temporal sequence segments of primary parameter encoding and apply these insights to shorten an exemplary MRF sequence by truncation.
|
|
| 16:57 | 0563.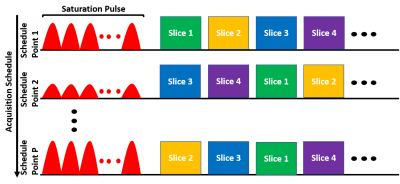 |
Fast CEST MR Fingerprinting with Increased Volumetric Coverage using Slice Permuted Acquisition and Deep Learning Reconstruction
Ouri Cohen1, Robert Young2, Christian T Farrar3, and Ricardo Otazo1
1Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3Radiology, Athinoula A. Martinos Center, Charlestown, MA, United States
CEST imaging is a promising tool for diagnosis and evaluation of treatment response in tumors. However, conventional CEST is not quantitative and requires long acquisition times. A recently developed technique, CEST MR fingerprinting (CEST-MRF), overcomes many of the technical limitations of conventional CEST but still suffers from limited volumetric coverage. In this work, we propose a novel multi-slice CEST-MRF pulse sequence and deep learning reconstruction method to enable volumetric coverage without the need for additional scan time. Numerical simulations and in vivo experiments in a healthy subject are performed to demonstrate feasibility and utility of the proposed multi-slice CEST-MRF technique.
|
|
| 16:59 | 0564.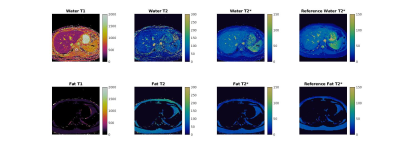 |
Simultaneous mapping of T1, T2, and T2* at 0.55T with Rosette MR Fingerprinting
Evan Cummings1,2, Yuchi Liu2, Yun Jiang1,2, Kathleen Ropella-Panagis2, Jesse Hamilton1,2, and Nicole Seiberlich1,2
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 2Radiology, University of Michigan, Ann Arbor, MI, United States
In this work, Rosette MRF was modified and deployed to simultaneously map T1, T2, and T2*. Mapping is demonstrated on the ISMRM/NIST phantom for verification of quantitative accuracy, and on four healthy volunteers to demonstrate potential feasibility in vivo.
|
|
| 17:01 | 0565.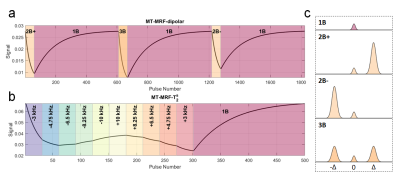 |
Towards quantification of semisolid tissue properties at 7T using multiband-MRF methods.
Daniel West1,2, Raphael Tomi-Tricot3, Lucilio Cordero-Grande1,4,5, Pip Bridgen1, Tom Wilkinson1, Sharon Giles1, Joseph Hajnal1,4, and Shaihan Malik1,4
1Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2London Collaborative Ultra High Field System (LoCUS), London, United Kingdom, 3MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom, 4Centre for the Developing Brain, King's College London, London, United Kingdom, 5Biomedical Image Technologies, ETSI Telecomunicación, Universidad Politécnica de Madrid & CIBER-BNN, Madrid, Spain
Since semisolid tissue components are strong determinants of relaxation parameters, estimation of semisolid properties may lead to more robust and direct characterization of tissues than relaxation measurement. We have used multiband MR fingerprinting at ultra-high field in an exploratory study towards high resolution quantitative imaging with efficient parameter encoding. To achieve this, two variants of a steady-state sequence are proposed that employ different cycling of nonselective multiband RF pulses: one to highlight dipolar order effects apparent in lipid bilayer-like materials such as myelin, and the other to measure semisolid T2. Initial phantom results and preliminary in vivo investigation are presented.
|
|
| 17:03 | 0566.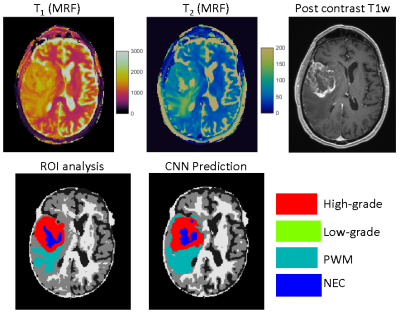 |
Tissue classification of cerebral gliomas using MR fingerprinting signal and deep learning
Yong Chen1, Rasim Boyacioglu1, Gamage Sugandima Nishadi Weragoda2, Michael Martens2, Mark Griswold1, and Chaitra Badve1,3
1Radiology, Case Western Reserve University, Cleveland, OH, United States, 2Physics, Case Western Reserve University, Cleveland, OH, United States, 3Radiology, University Hospitals Cleveland Medical Center, Cleveland, OH, United States
In this pilot study, we aim to analyze MR Fingerprinting (MRF) signal using deep learning network to assess the performance of tissue classification in gliomas. A U-Net based convolutional neural network was trained to learn glioma grades based on the SVD-compressed fingerprint acquired using MRF. Based on data acquired from a 5-minute MRF scan, the method shows great potential to accurately classify glioma grades without the need of image registration and contrast administration.
|
|
| 17:05 | 0567.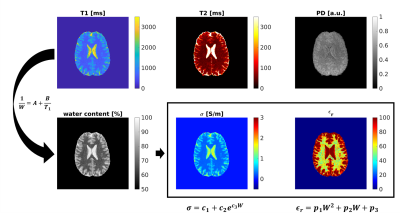 |
Fast high-resolution Electric Properties Mapping using three-dimensional MR Fingerprinting based water fraction estimation (MRF-EPT)
Matteo Cencini1,2, Marta Lancione1,2, Laura Biagi1,2, Rossella Pasquariello1, Luca Peretti2,3, Carolin M. Pirkl4, Rolf F. Schulte4, Guido Buonincontri1, Alessandro Arduino5, Luca Zilberti5, and Michela Tosetti1,2
1IRCCS Stella Maris, Pisa, Italy, 2Imago7 Foundation, Pisa, Italy, 3Università di Pisa, Pisa, Italy, 4GE Healthcare, Munich, Germany, 5Istituto Nazionale di Ricerca Metrologica (INRiM), Torino, Italy
MR-EPT aims to non-invasively assess the electrical properties (EP) of the tissues. However, conventional methods based on second-order derivatives of the transmit field are hampered by high noise amplification and boundary errors. Here, we propose to rely on the correlation between electrical properties and water content, which in turn is correlated to tissue T1, in order to extract EP maps from MR Fingerprinting based T1 estimation. We demonstrate whole-brain high-resolution EP mapping of healthy human volunteers and consistent results with both theoretical predictions and standard MR-EPT estimations.
|
|
| 17:07 | 0568.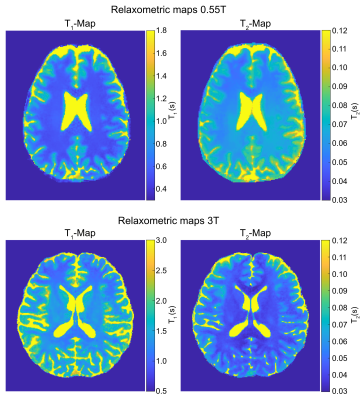 |
On the Feasibility of Hybrid-State based Quantitative MRI at 0.55T
Sebastian Flassbeck1,2 and Jakob Assländer1,2
1New York University School of Medicine, Center for Biomedical Imaging, New York, NY, United States, 2Center for Advanced Imaging Innovation and Research, New York University School of Medicine, New York, NY, United States
Low field magnetic resonance systems have potential as powerful diagnostic tools at comparably low cost. However, the inherently lower signal requires the use of SNR-efficient pulse sequences and reconstruction schemes. In this work, we present initial results with a hybrid-state free precession sequence, used to quantify T1 and T2 in 12min with an isotropic resolution of 1mm3. Visually, these quantitative maps show low noise levels, which allows for compelling synthetic MP-RAGE contrast generation.
|
|
| 17:09 | 0569.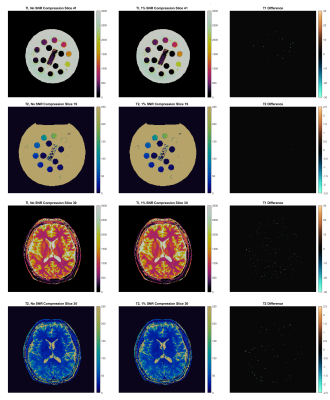 |
Online Gadgetron Reconstruction of 3D Magnetic Resonance Fingerprinting via a GPU-Accelerated Azure Kubernetes Cluster
Andrew Dupuis1, Yong Chen1, Rasim Boyacioglu1, John Stairs2, Michael Hansen2, Kelvin Chow3, and Mark A Griswold1
1Case Western Reserve University, Cleveland, OH, United States, 2Microsoft, Redmond, WA, United States, 3Siemens Medical Solutions USA, Inc., Chicago, IL, United States
Reconstruction of 3D Magnetic Resonance Fingerprinting acquisitions is computationally demanding, resulting in long processing times. GPU parallelization of the reconstruction’s NUFFT, pattern matching, and coil combination steps improves performance, but traditionally requires high-performance computers at the scanner. We propose an online reconstruction on a remote GPU-accelerated Kubernetes cluster. This allows many scanners or sites to share easily upgradeable and manageable computing resources. Additional calibration measurements, such as B1 maps, can also be transferred to allow inline B1 inhomogeneity correction. We also demonstrate that 3D-MRF reconstruction is robust with raw data compression that can be used to reduce site-to-cloud bandwidth requirements.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.