Power Pitch
Pitch: Deep/Machine Learning-Based Image Analysis & Application
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

Power Pitch Session: How it Works
1st Hour: 2-minute Power Pitches in the Power Pitch Theater.
2nd Hour: 60-minute digital poster presentations at the smaller screens around the perimeter of the Power Pitch Theater.
| 17:00 | 0167.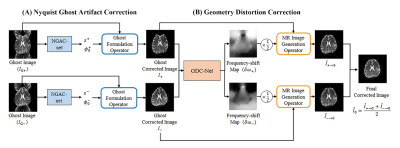 |
Unsupervised correction network for Nyquist ghost artifact and geometry distortion in echo planar imaging
Jeewon Kim1, Kinam Kwon1, Seohee So2, Byungjai Kim1, Wonil Lee1, and HyunWook Park1
1Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea, Republic of, 2Korea Institute of Science and Technology, Seoul, Korea, Republic of
We propose a new correction scheme using a deep neural network with unsupervised learning to correct Nyquist ghosts and geometry distortions occurring in EPI. The proposed scheme includes NGAC-net and GDC-net. First, the NGAC-net estimates the phase error of k-space with the help of a ghost formulation operator and correlation loss. The NGAC-net produces two Nyquist ghost corrected images obtained by dual-polarity phase-encoding gradients. The GDC-net is trained to estimate the frequency-shift map using the two output images from the NGAC-net. Afterwards, an MR image generation operator utilizes the estimated frequency-shift map to obtain the geometry distortion corrected images.
|
|
| 17:02 | 0168.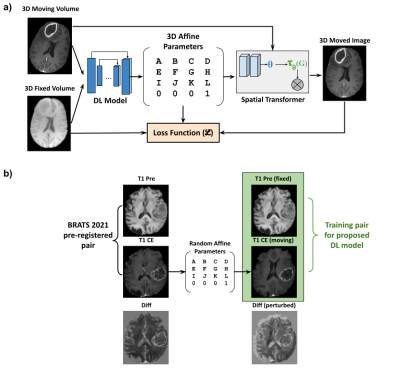 |
An unsupervised deep learning method for affine registration of multi-contrast brain MR images
Srivathsa Pasumarthi Venkata1, Ben Andrew Duffy1, and Keshav Datta1
1R&D, Subtle Medical Inc, Menlo Park, CA, United States
Image registration is a crucial preprocessing step for many downstream analysis tasks. Existing iterative methods for affine registration are accurate but time consuming. We propose a deep learning (DL) based unsupervised affine registration algorithm that executes orders of magnitude faster when compared to conventional registration toolkits. The proposed algorithm aligns 3D volumes from the same modality (e.g. T1 vs T1-CE) as well as different modalities (e.g. T1 vs T2). We train the model and perform quantitative evaluation using a pre-registered brain MRI public dataset.
|
|
| 17:04 | 0169.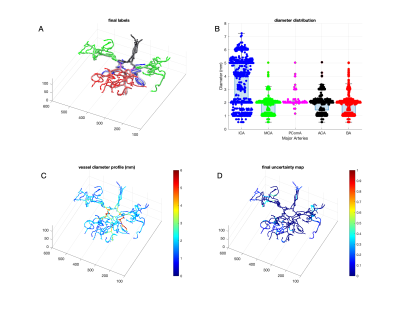 |
BayesTract: Automated machine learning based brain artery segmentation, anatomical prior annotation and feature-extraction in MR Angiography
Abrar Faiyaz1, Nhat Hoang1, Alan Finkelstein1, Jianhui Zhong1, Marvin Doyley1, Henry Wang2, Md Nasir Uddin1,2, and Giovanni Schifitto1,2
1University of Rochester, Rochester, NY, United States, 2University of Rochester Medical Center, Rochester, NY, United States Brain arterial-blood-vessels can carry important information regarding cerebrovascular pathogenesis. Due to non-invasiveness, 3D-time-of-flight MR-angiography is widely used to depict arteries in clinical-exams. However, deemed limited for qualitative assessment in clinical setting. Although several post-processing approaches exist such as tool-based-manual segmentation and diameter-marking or deep-learning-based segmentations--these require huge-time and experienced-eyes or large manually-segmented training-sets, making performance variable. Since brain-artery-annotations are anatomically-inspired, anatomical-confidant-points identified with a Bayesian approach can be used to annotate major-brain-arteries, which circumvents the need for large-dataset from learning requirement. The approach will allow clinical evaluations of major-arteries and biomarker identifications for cerebrovascular pathogenesis with limited resource and time. |
|
| 17:06 | 0170.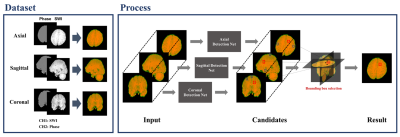 |
Triplanar Ensemble Detection Network (TPE-Det): A Single End-to-End Model for Efficient Detection of Cerebral Microbleeds in MR Images
Haejoon Lee1, Mohammed A. Al-masni1, Jun-Ho Kim1, Seul Lee1, Kyu-Jin Jung1, Woo-Ram Kim2, Young Noh2,3, and Dong-Hyun Kim1
1Department of Electrical & Electronic Engineering, Yonsei University, Seoul, Korea, Republic of, 2Neuroscience Research Institute, Gachon University, Incheon, Korea, Republic of, 3Department of Neurology, Gachon University College of Medicine, Gil Medical Center, Incheon, Korea, Republic of
Automated detection of Cerebral Microbleeds (CMBs) in Magnetic Resonance (MR) images can be clinically useful because of the small size of CMBs and the presence of numerous mimics. In this study, we propose an end-to-end Triplanar Ensemble Detection Network (TPE-Det) that demonstrates notable performance with only a single stage. Via the proposed TPE-Det, we combined CMBs detection results from axial, sagittal, and coronal planes. When considering the additional planes, the average False Positives per patient (FPavg) decreased from 29.16 to 0.92 compared to the case of using only the axial plane, preserving the high sensitivity of 95.94%.
|
|
| 17:08 | 0171.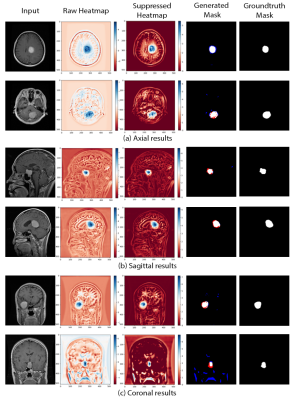 |
Learning to segment brain tumours using an explainable classifier
Soumick Chatterjee1,2,3, Hadya Yassin4, Florian Dubost5, Andreas Nürnberger2,3,6, and Oliver Speck1,6,7,8
1Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4Institute for Medical Engineerin, Otto von Guericke University Magdeburg, Magdeburg, Germany, 5Department of Biomedical Data Science, Stanford University, Stanford, CA, United States, 6Center for Behavioral Brain Sciences, Magdeburg, Germany, 7German Centre for NeurodegenerativeDiseases, Magdeburg, Germany, 8Leibniz Institute for Neurobiology, Magdeburg, Germany
Deep learning pipelines typically require manually annotated training data and the complex reasoning done by such methods make them appear as “black-boxes” to the end-users, leading to reduced trust. Unsupervised or weakly-supervised techniques could be a possible candidate for solving the first issue, while explainable classifiers or applying post-hoc interpretability-explainability methods on opaque classifiers may solve the second issue. This research addresses both problems by segmenting brain tumours without segmentation labels for training, using an explainable deep learning-based classifier. The classifier combined with a global pooling operation with a segmentation model to train and obtain classification results from this method.
|
|
| 17:10 | 0172.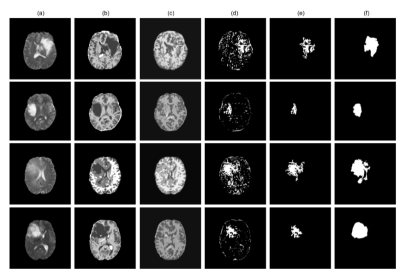 |
StRegA: Unsupervised Anomaly Detection in Brain MRIs using Compact Context-encoding Variational Autoencoder
Soumick Chatterjee1,2,3, Alessandro Sciarra1,4, Max Dünnwald3,4, Pavan Tummala3, Shubham Kumar Agrawal3, Aishwarya Jauhari3, Aman Kalra3, Steffen Oeltze-Jafra4,5, Oliver Speck1,5,6,7, and Andreas Nürnberger2,3,5
1Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4MedDigit, Department of Neurology, Medical Faculty, University Hospital, Magdeburg, Germany, 5Center for Behavioral Brain Sciences, Magdeburg, Germany, 6German Centre for NeurodegenerativeDiseases, Magdeburg, Germany, 7Leibniz Institute for Neurobiology, Magdeburg, Germany
Deep learning methods are typically trained in a supervised with annotated data for analysing medical images with the motivation of detecting pathologies. In the absence of manually annotated training data, unsupervised anomaly detection can be one of the possible solutions. This work proposes StRegA, an unsupervised anomaly detection pipeline based on a compact ceVAE and shows its applicability in detecting anomalies such as tumours in brain MRIs. The proposed pipeline achieved a Dice score of 0.642±0.101 while detecting tumours in T2w images of the BraTS dataset and 0.859±0.112 while detecting artificially induced anomalies.
|
|
| 17:12 | 0173.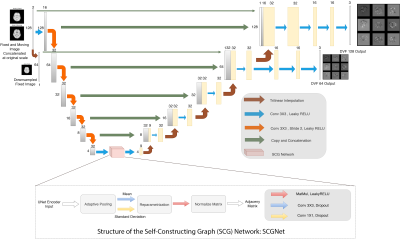 |
Multi-scale UNet with Self-Constructing Graph Latent for Deformable Image Registration
Soumick Chatterjee1,2,3, Himanshi Bajaj3, Mohammad Istiyak Hossain Siddiquee3, Nandish Bandi Subbarayappa3, Steve Simon3, Suraj Bangalore Shashidhar3, Oliver Speck1,4,5,6, and Andreas Nürnberger2,3,5
1Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4German Centre for NeurodegenerativeDiseases, Magdeburg, Germany, 5Center for Behavioral Brain Sciences, Magdeburg, Germany, 6Leibniz Institute for Neurobiology, Magdeburg, Germany
Deep Learning based deformable registration techniques such as Voxelmorph, ICNet, FIRE, do not explicitly encode global dependencies and track large deformations. This research attempts to encode semantics, i.e. structure and overall view of the anatomy in the supplied image, by incorporating self-constructing graph network in the latent space of a UNet model. It also attempts to track larger deformations through multiscale architecture and maintains consistent deformations through cycle consistency. The proposed method was compared against Voxelmorph and ANTs for T1 intramodal and T1-T2 Intermodal registration on IXI Dataset. The experiments show that the proposed model outperforms the baselines.
|
|
| 17:14 | 0174.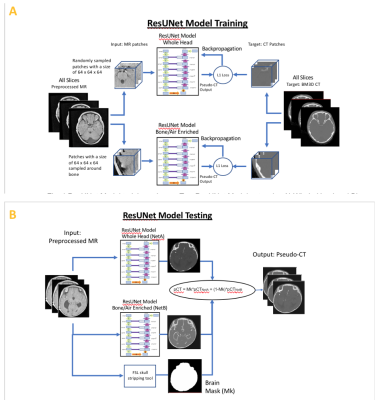 |
Deep learning-based high-resolution pseudo-CT to detect cranial bone abnormalities for pediatric patients using MRI
Parna Eshraghi Boroojeni1, Yasheng Chen1, Cihat Eldeniz1, Paul Commean1, Gary Skolnick1, Kamlesh Patel1, and Hongyu An1
1Washington University in Saint Louis, Saint Louis, MO, United States
Head trauma is common in the pediatric population. Craniosynostosis is abnormal early fusion of a cranial suture, causing an irregular-shaped cranium.3D high-resolution head CT scans are commonly used in these pediatric patients to identify skull fractures and sutures. However, CT exposespediatric patients to ionizing radiationand increases risk of cancer. We developed a robust and automated deep learning method to convert MR images to pseudo-CT (pCT) that can facilitate translating MR cranial bone imaging into clinical practice. An average Dice Coefficient of 0.89 and mean absolute error of 72.45 HU between pCT and CT were achieved.
|
|
| 17:16 | 0175.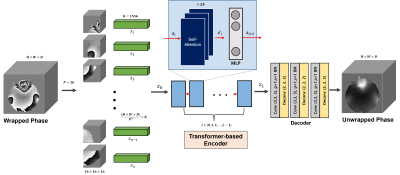 |
ViT-PU-Net: Volumetric Phase Unwrapping for MR images based on Vision Transformer
Soohyun Jeon1, Kanghyun Ryu2, and Dong-Hyun Kim1
1Electrical and Electronic Engineering, Yonsei University, Seoul, Korea, Republic of, 2Radiology, Stanford University, Stanford, CA, United States
MR phase information has been used in many MRI applications, recently, so it is very important to estimate the correct MR phase. Since the MR phase is encoded in complex exponential, the actual phase information obtained is in the form of a wrapped signal. Unfortunately, conventional MR phase unwrap methods are highly related to SNR and require a lot of computation, so an efficient method is needed.. Here, we propose ViT-PU-Net to perform 3D phase unwrap using ViT-based encoder architecture. Therefore, we investigated whether this architecture could be applied to 3D phase unwrap task by applying it to various data.
|
|
| 17:18 | 0176.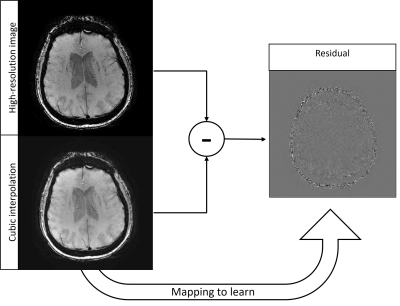 |
SWISeR: Multi-Field Susceptibility-Weighted Image Super-Resolution
Alexandra Grace Roberts1, Pascal Spincemaille2, Ilhami Kovanlikaya2, Apostolos John Tsiouris2, Thanh Nguyen2, and Yi Wang2,3
1Electrical and Computer Engineering, Cornell University, Ithaca, NY, United States, 2Radiology, Weill Cornell Medicine, New York, NY, United States, 3Biomedical Engineering, Cornell University, Ithaca, NY, United States
Susceptibility-Weighted Imaging Super-Resolution (SWISeR) increases the apparent resolution of input images and improves image quality, measured by mean-squared error and clinical scoring. This method generalizes to healthy subjects scanned at 3T and 7T and subjects imaged at 1.5T and 3T.
|
|
| 17:20 | 0177.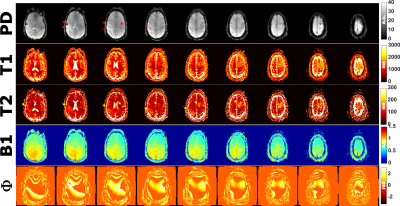 |
Rapid 3D Quantitative Mapping of Brain Metastases with Deep Learning-Based Phase-Sensitive MR fingerprinting
Victoria Y Yu1, Kathryn R Tringale2, Ricardo Otazo1, and Ouri Cohen1
1Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Radiation Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, United States
In MR fingerprinting, quantitative maps are obtained by matching the measured signal to a pre-computed dictionary. However, a key constraint of dictionary matching is the exponential growth of the dictionary with the number of parameters. A deep learning method named DRONE overcomes this constraint by using deep learning to map the magnitude-valued signal to the underlying tissue parameters. Here we describe an extension of DRONE that jointly estimates a phase term to enable mapping complex-valued signals and improve the quantitative accuracy. We test the accuracy in the ISMRM NIST phantom and demonstrate the clinical utility in patients with brain metastases.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.