Combined Educational & Scientific Session
Sharing Is Caring: Reproducible Research in MRI
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| 08:15 | Creating a Culture of Reproducible Research
Francesco Santini
Keywords: Transferable skills: Reproducible research The talk will present the importance of reproducibility in scientific research and how it has become a crucial part of scientific practices. The talk will also discuss the role of open data and source methods in promoting reproducibility and creating a more accessible and equitable scientific landscape. Finally, the talk will explain how implementing reproducible practices can improve the scientific output of labs in terms of both quality and efficiency. |
|
| 08:35 | Open-Source Hardware & Software Nikola Stikov | |
| 08:55 | 0396.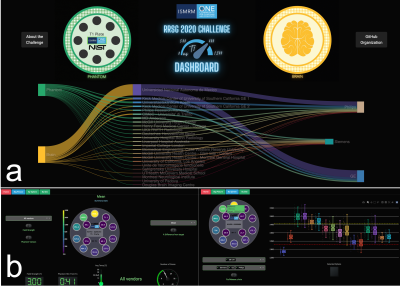 |
Phantom Results of the ISMRM Joint RRSG–qMRSG Reproducibility Challenge on T1 mapping
Mathieu Boudreau1,2, Agah Karakuzu1, Julien Cohen-Adad1,3, Madeline Carr4,5, Mariya Doneva6, Seraina A. Dual7,8, Daniel B. Ennis9, Alex Ensworth10,11, Alexandru Foias1, Véronique Fortier10,12, Guillaume Gilbert13, Matthew Grech-Sollars14,15, Lois Holloway4,5, Siyuan Hu16, Oscar Jalnefjord17,18, Peter Koken6, Anastasia Kolokotronis10,19, Simran Kukran20, Nam Lee21, Ives R. Levesque10,
Dan Ma16, Burkhard Maedler22, Nyasha Maforo23, Kévin Moulin9,24, Jamie Near25, Robba Rai4,5, Ben Statton26, Christian Stehning22, Chenyang Wang27, Kilian Weiss22, Niloufar Zakariaei28, Shuo Zhang22, and Nikola Stikov1,2
1NeuroPoly, Polytechnique Montreal, Montreal, QC, Canada, 2Montreal Heart Institute, Montreal, QC, Canada, 3Unité de Neuroimagerie Fonctionnelle (UNF), Centre de recherche de l’Institut Universitaire de Gériatrie de Montréal (CRIUGM), Montreal, QC, Canada, 4Medical Physics, Ingham Institute for Applied Medical Research, Liverpool, Australia, 5Department of Medical Physics, Liverpool and Macarthur Cancer Therapy Centres, Liverpool, Australia, 6Philips Research, Hamburg, Germany, 7Stanford University, Stanford, CA, United States, 8Department of Biomedical Engineering and Health Systems, KTH Royal Institute of Technology, Stockholm, Sweden, 9Department of Radiology, Stanford University, Stanford, CA, United States, 10Medical Physics Unit, McGill University, Montreal, QC, Canada, 11University of British Columbia, Vancouver, BC, Canada, 12Medical Imaging, McGill University Health Centre, Montreal, QC, Canada, 13MR Clinical Science, Philips Canada, Mississauga, ON, Canada, 14Department of Computer Science, University College London, London, United Kingdom, 15Lysholm Department of Neuroradiology, National Hospital for Neurology and Neurosurgery, University College London Hospitals NHS Foundation Trust, London, United Kingdom, 16Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 17Department of Medical Radiation Sciences, Institute of Clinical Sciences, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 18Department of Medical Physics and Biomedical Engineering, Sahlgrenska University Hospital, Gothenburg, Sweden, 19Hôpital Maisonneuve-Rosemont, Montreal, QC, Canada, 20Department of Surgery & Cancer, Imperial College London, London, United Kingdom, 21Department of Biomedical Engineering, Viterbi School of Engineering, University of Southern California, Los Angeles, CA, United States, 22Philips GmbH Market DACH, Hamburg, Germany, 23Philips Healthcare Germany, Hamburg, Germany, 24CREATIS Laboratory, Univ. Lyon, UJM-Saint-Etienne, INSA, CNRS UMR 5520, INSERM, Lyon, France, 25Centre d'Imagerie Cérébrale, Douglas Mental Health University Institute, Montreal, QC, Canada, 26MRC, London Institute of Medical Sciences, Imperial College London., London, United Kingdom, 27Department of Radiation Oncology - CNS Service, The University of Texas MD Anderson Cancer Center, Houston, TX, United States, 28Department of Radiation Oncology, Henry Ford Cancer Institute, Detroit, MI, United States Keywords: Quantitative Imaging, Relaxometry, Reproducibility, challenge A collaborative reproducibility challenge was launched to explore if an imaging protocol independently-implemented at multiple centers can reliably measure T1 using inversion recovery in a standardized quantitative MRI phantom (ISMRM/NIST). A total of 19 submissions were accepted, totalling 41 phantom T1 mapping datasets. Errors relative to the temperature-corrected reference T1 values were under 10% for the range of values expected in the human brain in vivo. All submitted phantom data, code, pipelines, and scripts were shared on open platforms. |
09:03 |
0397.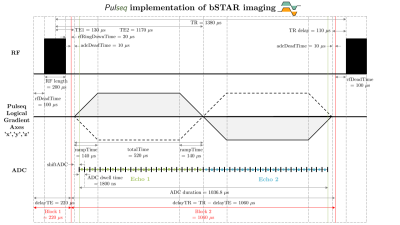 |
Reproducibility of the bSTAR sequence and open-source implementation
Nam G Lee1, Grzegorz Bauman2, Oliver Bieri2, and Krishna S Nayak3
1Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 2Department of Radiology, University of Basel Hospital, Basel, Switzerland, 3Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States Keywords: Lung, Low-Field MRI The reproducibility of scientific reports is crucial to advancing human knowledge. This abstract is a response to the 2023 ISMRM Challenge “Repeat it With Me: Reproducibility Team Challenge”. We reproduce the bSTAR sequence, a very short-TR, 3D half-radial dual-echo bSSFP sequence, providing banding-artifact free images within a large FOV. bSTAR imaging is attractive for various applications at low field and especially attractive for lung parenchyma imaging due to the prolonged T2’. We have successfully reproduced the bSTAR method, and figures with comparable image quality compared to published literature. We provide an open-source implementation using Pulseq and BART. |
09:11 |
0398.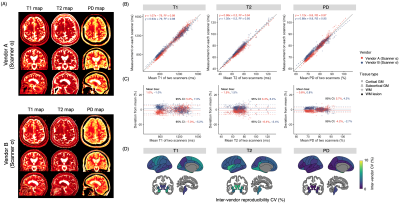 |
Cross-vendor three-dimensional multiparametric mapping of the human brain: A traveling-subject and patient study
Shohei Fujita1,2, Borjan Gagoski3,4, Ken-Pin Hwang5, Marcel Warntjes6,7, Kazumasa Yokoyama8,9, Issei Fukunaga1, Wataru Uchida1, Yuya Saito1, Rina Tachibana1, Tomoya Muroi1, Toshiya Akatsu1, Akihiro Kasahara2, Ryo Sato2, Tsuyoshi Ueyama2, Christina Andica1, Akifumi Hagiwara1, Koji Kamagata1, Shiori Amemiya2, Hidemasa Takao2, Nobutaka Hattori9,
Osamu Abe2, and Shigeki Aoki1
1Department of Radiology, Juntendo University, Tokyo, Japan, 2Department of Radiology, The University of Tokyo, Tokyo, Japan, 3Fetal Neonatal Neuroimaging and Developmental Science Center, Boston Children's Hospital, Boston, MA, United States, 4Department of Radiology, Harvard Medical School, Boston, MA, United States, 5Department of Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States, 6SyntheticMR, Linköping, Sweden, 7Center for Medical Imaging Science and Visualization (CMIV), Linköping University, Linköping, Sweden, 8Tousei center for neurological diseases, Shizuoka, Japan, 9Department of Neurology, Juntendo University, Tokyo, Japan Keywords: Quantitative Imaging, Precision & Accuracy, Cross-vendor Multiparametric techniques compatible with multiple vendors to facilitate the pooling of data among different sites and vendors are desired. Here, we developed a vendor-standardized whole-brain multiparametric mapping scheme based on 3D-QALAS. Intra-scanner repeatability and inter-vendor reproducibility were evaluated on test-retest session data on five different 3T systems from four MRI vendors (GE, Philips, Siemens, and Canon). T1 and T2 relaxation times and proton density values derived from 3D-QALAS showed coefficient of variations of <4.0% across scanners from different vendors. Finally, we performed an inter-vendor validation on multiple sclerosis patients to assess the feasibility of the scheme in real-world clinical settings. |
| 09:19 | 0399. |
A Multi-Site Collaborative Training Effort to Improve Neuroimaging Accessibility and Capacity Development in Low- and Middle-Income Countries
Jessica E. Ringshaw1,2,3, Layla E. Bradford1,2, Marlie Miles1,2, Carly Bennallick3, Niall J. Bourke3, Emil Ljungberg3,4, Sean C.L. Deoni5, Steven C.R. Williams3, and Kirsten A. Donald1,2
1Department of Paediatrics and Child Health, University of Cape Town, Cape Town, South Africa, 2Neuroscience Institute, University of Cape Town, Cape Town, South Africa, 3Department of Neuroimaging, King's College London, London, United Kingdom, 4Department of Medical Radiation Physics, Lund University, Lund, Sweden, 5Maternal, Newborn, and Child Health (MNCH) Discovery and Tools (D&T), Bill and Melinda Gates Foundation, Seattle, WA, United States Keywords: Neuro, Low-Field MRI, Training, Capacity Development, Low- and Middle Income Countries In a multi-site global collaboration (UNITY; Ultra-Low Field Neuroimaging In The Young) implementing the novel Hyperfine 64mT low-field MRI into research on relevant health priorities in low- and middle-income countries (LMICs), capacity development has been identified as a key objective. Following an initial training wave in foundational sites including South Africa, adapted workshops have been conducted in new consortium countries including Malawi, Ethiopia, and Ghana. This has informed the development of a strategic framework for context-specific training aimed at promoting the increased success of neuroimaging in research and clinical practice, and the sustainability of MRI technology in under-resourced settings. |
| 09:27 | 0400.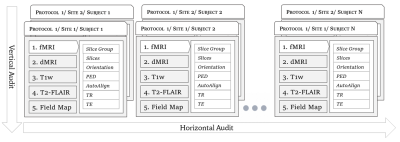 |
Solving the pervasive problem of protocol non-compliance in MR imaging using an open-source tool mrQA
Harsh Sinha1 and Pradeep Reddy Raamana2
1Intelligent Systems Program, University of Pittsburgh, Pittsburgh, PA, United States, 2Intelligent Systems Program, Department of Biomedical Informatics, Department of Radiology, University of Pittsburgh, Pittsburgh, PA, United States Keywords: Data Acquisition, Data Processing, Protocol Compliance, Quality Control, Quality Assurance Acquiring MR imaging data by multi-site consortia requires careful monitoring of MR physics protocols. Conventionally, protocol compliance has been an ad-hoc and manual process that is error-prone. Hence, it is often overlooked for the lack of realization that parameters are routinely improvised locally at different sites. Such variation and inconsistencies across acquisition protocols can reduce SNR, & statistical power and, in the worst case, may invalidate the results altogether. We present an open-source tool, called mrQA, to assess protocol compliance and demonstrate the lack of it by analyzing over 20 large open datasets. |
| 09:35 | 0401.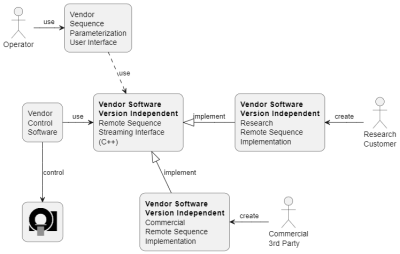 |
A Remote Sequence Streaming Interface for Open Innovation
Thomas Kluge1 and Christoph Forman1
1Siemens Healthcare GmbH, Erlangen, Germany Keywords: Data Acquisition, Pulse Sequence Design, Open Innovation; Reproducible Research Open innovation and reproducible research have become increasingly important in recent years. Furthermore, software version dependent application development introduces significant efforts to developers and researchers in maintaining different versions of applications for a variety of scanner software releases. In this work, we propose a software-version and programming language independent interface for data acquisition on Siemens Healthcare MAGNETOM scanners. This is achieved by generic interfaces and streaming protocols. A wide range of development and deployment opportunities is enabled with a web-API. The use of the interface is exemplarily shown with a prototype FLASH sequence and demonstrated with a phantom scan. |
| 09:43 | 0402.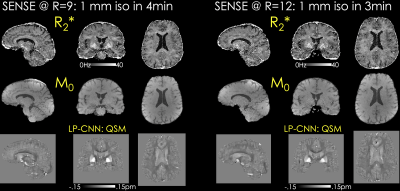 |
An Open-Source Self-navigated Multi-Echo Gradient Echo Acquisition for R2* and QSM mapping using Pulseq and Model-Based Reconstruction
Xiaoqing Wang1,2, Berkin Bilgic1,2, Daniel Gallichan3, Kwok-Shing Chan4, and José P. Marques4
1Massachusetts General Hospital, Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Cardiff University Brain Research Imaging Centre, Cardiff, United Kingdom, 4Donders Institute for Brain, Cognition and Behavior, Radboud University, Nijmegen, Netherlands Keywords: Pulse Sequence Design, Quantitative Susceptibility mapping, open-source, pulseq We propose an open-source multi-echo GRE sequence and reconstruction algorithms for rapid acquisition and harmonization of QSM. This is implemented using Pulseq, where different sampling patterns are used across echoes to provide complementary information and facilitate the proposed MOdel BAsed (MOBA) reconstruction. MOBA estimates M0, R2* and frequency maps using nonlinear optimization and is implemented in BART to facilitate dissemination. B0 correction is performed using 1D navigators acquired during the crusher gradients in each repetition. Motion navigators inserted in the sequence permit robust estimation of motion parameters with time frames of ~13seconds, and will lend themselves to retrospective motion correction. |
| 09:51 | 0403.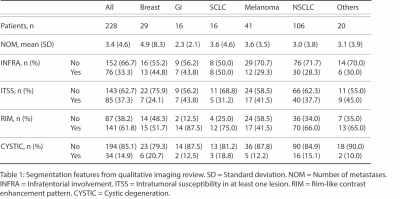 |
Developing an Open Access Brain Metastasis Database: Yale Brain Metastasis Database
Divya Ramakrishnan1, Leon Jekel2, Matthew Sala1, Manpreet Kaur3, Anastasia Janas1, Gabriel Cassinelli Petersen4, Khaled Bousabarah5, MingDe Lin6, Sara Merkaj7, Marc von Reppert8, and Mariam Aboian1
1Department of Radiology, Yale School of Medicine, New Haven, CT, United States, 2University of Essen, Essen, Germany, 3Ludwig Maximilians Universität (LMU), Munich, Germany, 4University of Goettingen, Goettingen, Germany, 5Visage Imaging, Dusseldorf, Germany, 6Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States, 7University of Ulm, Ulm, Germany, 8University of Leipzig, Leipzig, Germany Keywords: Tumors, Machine Learning/Artificial Intelligence, Brain Metastasis While there are many machine learning (ML) algorithms for brain metastasis (BM) detection and segmentation, very few have been validated on external datasets. There is a critical need for open access BM datasets for development and validation of more robust algorithms. Here, we present the Yale Brain Metastasis database of 290 patients with annotated segmentations of BM on T1 post-gadolinium and associated survival information. A subset of 228 patients have FLAIR segmentations, clinical features, and qualitative imaging features. Open access of this database will greatly aid in the development and validation of new AI algorithms for BM detection and segmentation. |
| 09:59 | 0404.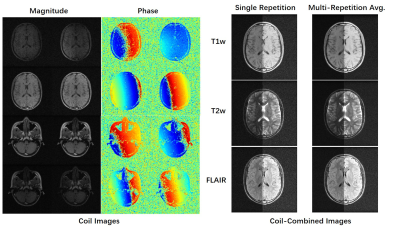 |
M4Raw: A Multi-Contrast Multi-Repetition Multi-Channel Raw K-space Dataset for Low-Field MRI Reconstruction
Mengye Lyu1, Lifeng Mei1, Sixing Liu1, Shoujin Huang1, Yi Li1, Kexin Yang1, Yilong Liu2, and Ed X. Wu3,4
1College of Health Science and Environmental Engineering, Shenzhen Technology University, Shenzhen, China, 2Guangdong-Hongkong-Macau Institute of CNS Regeneration, Key Laboratory of CNS Regeneration (Ministry of Education), Jinan University, Guangzhou, China, 3Laboratory of Biomedical Imaging and Signal Processing, the University of Hong Kong, Hong Kong, China, 4Department of Electrical and Electronic Engineering, the University of Hong Kong, Hong Kong, China Keywords: Machine Learning/Artificial Intelligence, Low-Field MRI, Open dataset We release a new raw k-space dataset M4Raw acquired by the low-field MRI. Currently, it contains multi-channel brain data of 180 subjects each with 18 slices x 3 contrasts (T1w, T2w, and FLAIR). Moreover, each contrast consists of two or three repetitions (a.k.a. NEXs), leading to more than 25k trainable slices in total, which can be used in various ways by the low-field MRI community. It can be accessed via http://github.com/mylyu/M4Raw. |
| 10:07 | 0405.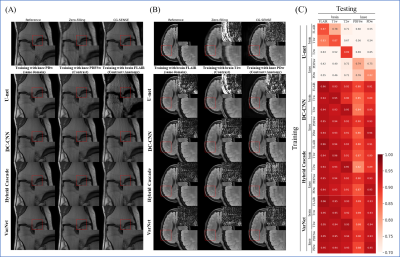 |
Systematic evaluation of the robustness of open-source networks for MRI multicoil reconstruction
Naoto Fujita1, Suguru Yokosawa2, Toru Shirai2, and Yasuhiko Terada1
1Graduate School of Science and Technology, University of Tsukuba, Tsukuba, Japan, 2FUJIFILM Healthcare Corporation, Tokyo, Japan Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, Deep Learning Reconstrcution Deep neural networks (DNNs) for MRI reconstruction often require large datasets for training, but in clinical settings, the domains of datasets are diverse, and the degree to which deep neural networks are the robustness of DNNs to domain differences between training and testing datasets has been an open question. Here, we evaluated the robustness of four open-source multicoil networks to differences in the domain. We found that model-based networks exhibit higher robustness than data-driven networks and that robustness varies across network architectures, even within model-based networks. Our results provide insight into what network architectures are effective for generalization performance. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.