Digital Poster
Segmentation II
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
3760.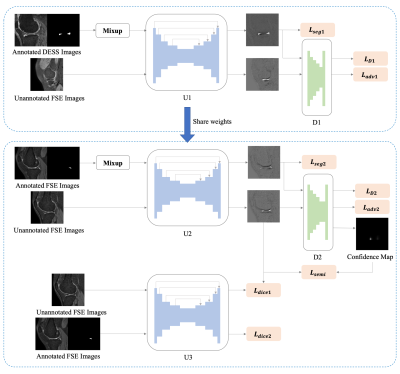 |
1 |
A deep neural network framework of few-shot learning with
domain adaptation for automatic meniscus segmentation in 3D Fast
Spin Echo MRI
Siyue Li1,
Shutian Zhao1,
Fan Xiao2,
Dόnal G. Cahil3,
Kevin Ki-Wai Ho4,
Michael Tim-Yin Ong4,
Queeie Chan5,
James F Griffith3,
Jin Hong6,
and Weitian Chen1
1CU Lab for AI in Radiology (CLAIR), Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, China, 2Department of Radiology, Shanghai Sixth People's Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shang hai, China, 3Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, China, 4Department of Orthopaedics & Traumatology, The Chinese University of Hong Kong, Hong Kong, China, 5Philips Healthcare, Hong Kong, China, 6Guangdong Provincial People's Hospital, Guangdong Academy of Medical Sciences, Guang Zhou, China Keywords: Segmentation, Machine Learning/Artificial Intelligence Automatic meniscus segmentation is highly desirable for quantitative analysis of knee joint diseases. As three-dimensional Fast Spin Echo (3D FSE) is a promising MR (magnetic resonance) imaging technique to evaluate the tissues of the knee joint. In this study, we explore meniscal segmentation on 3D FSE MRI. Manually annotating 3D knee images is challenging since it is time-consuming and requires clinical expertise. In this study, we propose a domain adaption-based few-shot learning method for meniscal segmentation on 3D FSE images using only one annotated MRI data. We demonstrate that the proposed method outperformed the fully supervised segmentation model. |
|
3761.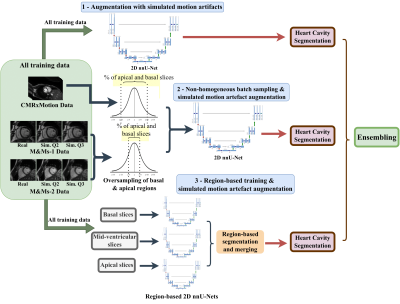 |
2 |
Cardiac MR Image Segmentation in the Presence of Respiratory
Motion Artifacts
Yasmina Al Khalil1,
Sina Amirrajab1,
Josien Pluim1,
Marcel Breeuwer1,2,
and Cian Scannell1
1Biomedical Engineering Department, Eindhoven University of Technology, Eindhoven, Netherlands, 2MR R&D - Clinical Science, Philips Healthcare, Best, Netherlands Keywords: Segmentation, Segmentation Object motion during the acquisition of magnetic resonance images can negatively impact image quality by introducing inconsistencies in the k-space data and in turn, blurring and ghosting artifacts on the acquired images. Such artifacts represent significant challenges in the clinical deployment of automated segmentation algorithms. In this work, we present an ensemble of approaches aimed at developing a robust and generalizable segmentation model, particularly tailored to handle the appearance of respiratory motion artifacts. We achieve this by introducing k-space based simulation for augmentation, as well as by reducing common errors across basal and apical slices with a region-focused segmentation approach.
|
|
3762. |
3 |
A-Eye: Towards a large-scale MRI-based model of the complete eye
Jaime Barranco1,2,
Hamza Kebiri1,2,
Óscar Esteban3,
Raphael Sznitman4,
Oliver Stachs5,
Sönke Langner6,7,
Benedetta Franceschiello8,9,10,
and Meritxell Bach Cuadra1,2,10
1CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 2Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 4ARTORG Center for Biomedical Engineering, University of Bern, Bern, Switzerland, 5Ophthalmology, Rostock University Medical Center, Rostock, Germany, 6Institute for Diagnostic and Interventional Radiology, Rostock University Medical Center, Rostock, Germany, 7Diagnostic Radiology and Neuroradiology, University of Greifswald, Greifswald, Germany, 8School of Engineering, Institute of Systems Engineering, HES-SO Valais-Wallis, Sion, Switzerland, 9The Sense Innovation and Research Center, Lausanne and Sion, Switzerland, 10These authors provided equal last-authorship contribution, Lausanne, Switzerland Keywords: Segmentation, Neuro, Eye, ophthalmology This work comparatively evaluates two approaches for the automated segmentation of eye structures from 3D T1-weighted MRI data of the whole human head (N=1210). Quantitative results on a validation sub-set with manual annotations provide accurate results for lens and globe and set the first median Dice (DSC) benchmarks for optic-nerve (0.91), muscles (0.58 to 0.76) and fat (0.67 and 0.75). The ability of our framework to automatically extract state-of-the-art measurements, such as the axial length, paves the way to accurately identify and compute new biomarkers of the eye via MRI. |
|
3763.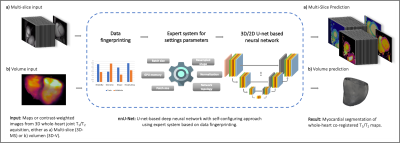 |
4 |
Automatic segmentation of myocardial 3D whole-heart T1 and T2
maps using a nnU-Net.
Carlos Velasco1,
Roman Jakubicek2,
Alina Hua1,
Anastasia Fotaki1,
René M. Botnar1,3,
and Claudia Prieto1,3
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Department of Biomedical Engineering, Brno University of Technology, Brno, Czech Republic, 3Institute for Biological and Medical Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile Keywords: Segmentation, Myocardium The high amount of data obtained from a single 3D whole heart multiparametric scan (up to ~40 slices per parametric map) increases considerably the time required to segment and analyse the quantitative maps. Thus, an automated segmentation tool for these maps is desirable to perform this otherwise prohibitively laborious task. In this work, we leverage the potential of nnU-Net to perform fast, automated segmentation of 3D whole-heart simultaneous T1 and T2 maps and show its feasibility to predict segmentation masks with comparable quality while shortening the segmentation and analysis time by ~100x. |
|
3764.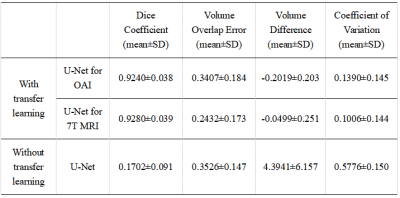 |
5 |
Automated Segmentation of Knee Cartilage from Ultra-High
Resolution 7 Tesla 3D bSSFP MRI Using Transfer Learning
Luxuan Guo1,
Simran Kukran1,
Krithika Balaji1,
and Neal Bangerter1
1Imperial College London, London, United Kingdom Keywords: Segmentation, Cartilage, Osteoarthritis 7T 3D knee MRI shows great promise for the quantification of cartilage volume and thickness to assess osteoarthritis, but manual segmentation is time consuming. Segmented 7T 3D knee datasets to train machine learning techniques are limited. Here, we trained a network on a larger dataset of segmented 3T 3D knee MRI scans and used transfer learning to create an automatic segmentation network of 7T MRI knee cartilage using a small number of segmented 7T images. The resulting network constructed demonstrated vastly improved automatic segmentation of the knee cartilage at 7T compared to the network trained with limited 7T data only. |
|
3765.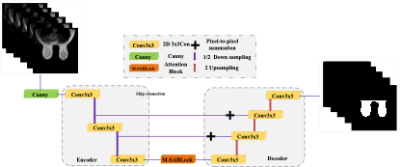 |
6 |
Breast Segmentation of MRI Based on U-Net and Multi-Headed
Self-Attention Mechanism
Hang Yu1,
Yuru Guo1,
Lizhi Xie2,
Zhiheng Liu1,
Zichuan Xie3,
Chenyang Li1,
and Suiping Zhou1
1School of Aerospace Science and Technology,Xidian university, xi'an, China, 2GE Healthcare, Beijing, China, 3Guangzhou institute of technology,Xidian University, Guangzhou, China Keywords: Segmentation, Breast In breast MRI, overall breast segmentation is a key step in performing breast cancer risk assessment.To achieve automatic and accurate breast segmentation in breast MR images, we propose a breast segmentation model based on U-Net and multi-head self-attention mechanism, which adds global information through multi-head self-attention module and changes the cascade structure in U-Net network to pixel-by-pixel summation.On the breast MRI dataset, the proposed model can achieve accurate, effective and fast breast segmentation with an average DSC and an average MIOU of 97.28 % and 92.01 %, respectively, which are 4.7 % and 5.56 % higher compared to U-Net, respectively. |
|
3766.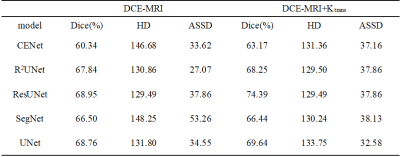 |
7 |
The effectiveness of the volume transfer constant (Ktrans) as an
aid to regional segmentation of nasopharyngeal tumors
Junhui Huang1,2,
Zhou Liu3,
Liyan Zou3,
Yingying Chen3,
Zhanli Hu4,
Dong Liang4,
Xin Liu4,
Hairong Zheng4,
and Na Zhang4
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, ShenZhen, China, 2Faculty of Information Engineering and Automation, Kunming University of Science and Technology, Kunming, China, 3Department of Radiology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen, China, 4Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China Keywords: Segmentation, Machine Learning/Artificial Intelligence Accurate segmentation of nasopharyngeal tumor lesions from dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) facilitates subsequent diagnosis and treatment. However, current segmentation methods do not incorporate the pathological properties of the tumor. Therefore, this paper proposes a multimodal DCE-MRI segmentation method that uses the pharmacokinetic features of NPC, Ktrans, as modal information to assist nasopharyngeal tumor segmentation. We validated our method in several classical deep learning segmentation networks, and DCE-MRI with fused Ktrans eigenmodes had higher Dice coefficients than DCE-MRI with a single modality. The best segmentation results were obtained by this method on the ResUNet model (dice=74.39). |
|
3767.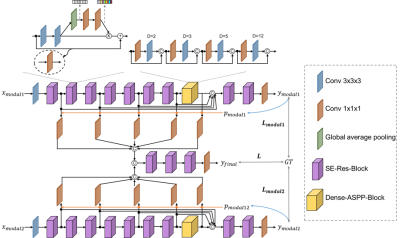 |
8 |
Complementarity-aware multi-parametric MR image feature fusion
for abdominal multi-organ segmentation
Cheng Li1,2,
Yousuf Babiker M. Osman1,3,
Weijian Huang1,3,4,
Zhenzhen Xue1,2,
Hua Han1,3,
Hairong Zheng1,
and Shanshan Wang1,2,4
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application, Guangzhou, China, 3University of Chinese Academy of Sciences, Beijing, China, 4Peng Cheng Laboratory, Shenzhen, China Keywords: Segmentation, Body T1-weighted in-phase and opposed-phase gradient-echo imaging is a routine component in abdominal MR imaging. Organ segmentation with the acquired images plays an important role in identifying various diseases and making treatment plans. Despite the promising performance achieved by existing deep learning models, further investigation is still needed to effectively exploit the information provided by different imaging parameters. Here, we propose a complementarity-aware multi-parametric MR image feature fusion network to extract and fuse the information of paired in-phase and opposed-phase MR images for enhanced abdominal multi-organ segmentation. Extensive experiments are conducted, and better results are achieved when compared to existing methods. |
|
3768.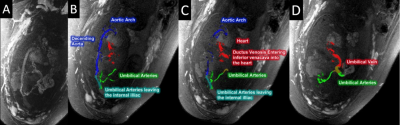 |
9 |
Extraction of the Utero-Placental and Fetal Vasculature using 2D
Time-of-Flight Imaging
Karthikeyan Subramanian1,
Pavan Kumar Jella1,
Feifei Qu1,
Tinnakorn Chaiworapongsa2,3,
and Mark E Haacke1
1Department of Radiology, Wayne State University, Detroit, MI, United States, 2Perinatology Research Branch, NICHD/NIH/DHHS, Detroit, MI, United States, 3Department of Obstetrics and Gynecology, Wayne State University, Detroit, MI, United States Keywords: Segmentation, Fetus Mapping the vasculature, flow and tissue properties of the placenta and umbilical cord can serve as a means to study placental and fetal health. The goal of this work is to use a rapid, multi-echo, interleaved GRE sequence to minimize motion artifacts and cover the entire abdomen of the mother in a few minutes. To better map out the vasculature, we propose to both separate the arteries and veins in the umbilical cord using an R2* mapping thresholding technique and use vessel tracking to create 3D renderings. We have successfully done this in a series of 10 fetuses. |
|
3769.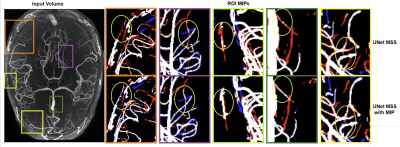 |
10 |
Enhancing Vessel Continuity in Deep Learning based Segmentation
using Maximum Intensity Projection as Loss
Soumick Chatterjee1,2,3,
Karthikesh Varma Chintalapati1,
Chethan Radhakrishna1,
Sri Chandana Hudukula Ram Kumar1,
Raviteja Sutrave1,
Hendrik Mattern3,
Oliver Speck3,4,5,
and Andreas Nürnberger1,2,5
1Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4German Center for Neurodegenerative Disease, Magdeburg, Germany, 5Center for Behavioral Brain Sciences, Magdeburg, Germany Keywords: Segmentation, Blood vessels Vessel Segmentation with deep learning is a challenging task that involves not only learning high-level feature representations but also the spatial continuity of the features across dimensions. Semi-supervised patch-based approaches have been effective in identifying small vessels of 1-2 voxels in diameter but failed to maintain vessel continuity. This study focuses on improving the segmentation quality by considering the spatial correlation of the features using the maximum intensity projection (MIP) as an additional loss criterion. It was observed that the proposed method quantitatively improves the segmentation while also improving vessel continuity, as evident in the visual examinations of ROIs. |
|
3770.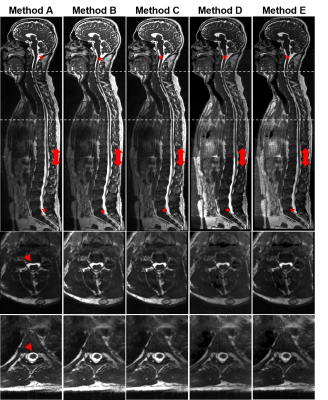 |
11 |
Full spine 3D T2-weighted MRI with improved stitching for
segmentation of the intrathecal space
Catarina Rua1,
Mari Lambrechts1,
Mark Tanner1,
James Davies1,
Ali Ghayoor2,
Howard Dobson2,
and Lino Becerra2
1Invicro LLC, A Konica Minolta Company, London, United Kingdom, 2Invicro LLC, A Konica Minolta Company, Needham, MA, United States Keywords: Segmentation, Neurofluids, CSF segmentation spine Computational fluid dynamics (CFD) has been used to model the behavior of cerebral-spinal-fluid (CSF) flow along the spine in research of intrathecal drug delivery. However, the model geometry highly impacts the CFD-based prediction of CSF flow. Enhanced images of the spinal CSF can be obtained with T2-weighted MRI, typically collected in 3-4 stations. Here we present a method to stitch spine images for CSF segmentation and compare it to the scanner’s stitching technique. Our offline approach appears more robust to movement and field-of-view adjustments while scanning and presents acceptable CSF signal intensity homogeneity across the spine for tissue segmentation. |
|
3771.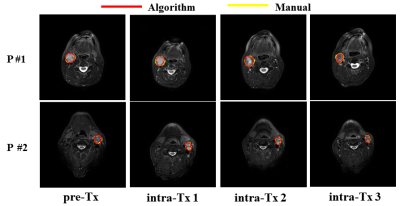 |
12 |
Deep learning-based auto-segmentation of neck nodal metastases
on longitudinal MR images using self-distilled masked image
transformer
Jue Jiang1,
Ramesh Paudyal1,
Bill H. Diplas2,
James Han2,
Nadeem Riaz2,
Vaios Hatzoglou 3,
Nancy Lee2,
Joseph Deasy 1,
Amita Shukla-Dave 1,3,
and Harini Veeraraghavan 1
1Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Radiation Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States Keywords: Segmentation, Cancer Manual segmentation of normal and tumor tissues on MRI is a traditional approach that is still used, but it is a very challenging and time-consuming method requiring a high level of precision and has shown inter-reader contouring variability. Therefore, semi- or fully automated segmentation algorithms are essential to segment tumors such as neck nodal metastases. The present study aimed to apply the previously developed deep learning-based self-distilled masked image transformer method for auto-segmenting neck nodal metastases on longitudinal T2-weighted MR images. |
|
3772.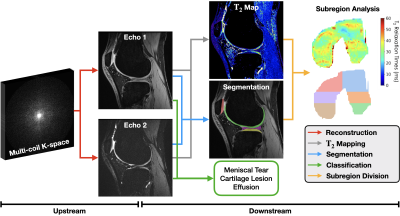 |
13 |
Assessing the Impact of Upstream Reconstruction Models on
Downstream Image Analysis: A Workflow-Centric Evaluation
Ben Viggiano1,
Aashna Desai2,
Elka Rubin3,
Andrew Schmidt3,
Robert Boutin3,
Kathryn J Stevens3,
Garry E Gold3,
Christopher Ré4,
Akshay S Chaudhari1,3,
and Arjun D Desai3,5
1Biomedical Data Science, Stanford University, Stanford, CA, United States, 2Department of Neuroscience, University of California Berkeley, Berkeley, CA, United States, 3Department of Radiology, Stanford University, Stanford, CA, United States, 4Department of Computer Science, Stanford University, Stanford, CA, United States, 5Department of Electrical Engineering, Stanford University, Stanford, CA, United States Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, Segmentation, Classification Deep learning (DL) techniques have shown promise for both reconstruction and image analysis stages of MRI workflows. However, traditional benchmarking methods evaluate each stage separately. As a result, the impact of reconstruction on downstream image analysis tasks and biomarker quantification remains unknown. In this study, we explore how changing aspects of upstream reconstruction affects the downstream analysis. We find that insights from evaluating reconstruction models as a component of a broader end-to-end workflow do not correlate with conventional, task-specific image quality metrics. We use these findings to motivate the discussion of evaluating DL methods at the workflow level. |
|
3773.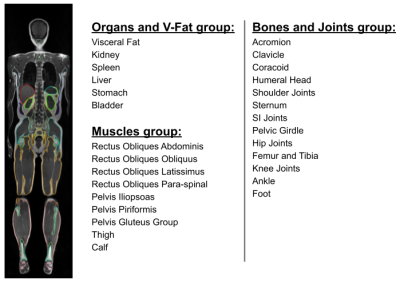 |
14 |
Age Related Changes of Organs and Muscles Using a Deep Learning
Based Whole Body Segmentation Technique Applied to a Healthy
Adult Population
Ahmed Gouda1,
Saqib Basar1,
Yosef Chodakiewitz2,
Rajpaul Attariwala1,
Sean London2,
and Sam Hashemi1
1Voxelwise Imaging Technology Inc., Vancouver, BC, Canada, 2Prenuvo, Vancouver, BC, Canada Keywords: Data Analysis, Aging, Volumetric Analysis, Organs, Visceral Fat, Skeletal Muscles One of the vital indicators of normal functioning physiology and anatomy, is the volume and size of body organs and muscles. Overtime due to aging and chronic illnesses, the volume of organs and muscles decreases, and quantifying this reduction would help us in predicting the rate of decline, and making efforts to improve this rate. Artificial intelligence 3D segmentation techniques allow us to measure the average age-related volume loss over a large population. They provide statistical norms which help radiologists to identify abnormal volume changes, and establish a normal age related rate of volumetric decline. |
|
3774. |
15 |
An Adaptive Weighted Active Contour Segmentation Model for 3T/5T
MRI from the Same Person
Zhenxing Huang1,
Mengxiao Geng1,
Liyun Zheng2,
Yongming Dai3,
Na Zhang1,
Dong Liang1,
Hairong Zheng1,
and Zhanli Hu1
1Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Shenzhen United Imaging Research Institute of Innovative Medical Equipment, Shenzhen, China, 3Central Reasearch Institute, United Imaging Healthcare, Shanghai, China Keywords: Data Analysis, Segmentation, 3T/5T MRI Image segmentation is a complex and core technique in the medical image domain. However, low-quality images, such as images with weak edges, may bring considerable challenges for radiologists. In this paper, we propose an adaptive weighted curvature-based active contour model by coupling heat kernel convolution and adaptively weighted high-order total variation to improve diagnosis effectiveness. The numerical experimental results on 3T/5T MRI datasets demonstrate that the proposed model is quite efficient and robust compared with several traditional segmentation methods, which would exert great value in quantitative image evaluation of MRI diagnosis for the same person. |
|
3775.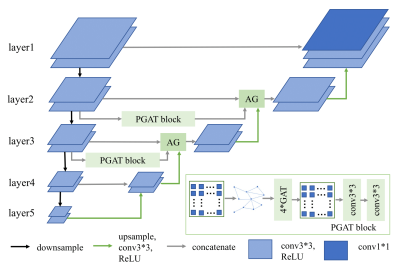 |
16 |
HYBRID MULTI-LEVEL GRAPH NEURAL NETWORK FOR CARDIAC MAGNETIC
RESONANCE SEGMENTATION
Xiaodi Li1,
Peng Li1,
and Yue Hu1
1Harbin Institute of Technology, Harbin, China Keywords: Data Processing, Segmentation Due to the inherent locality of convolutional operations, convolution neural network (CNN) often exhibits limitations in explicitly modeling long-distance dependencies. In this paper, we propose a novel hybrid multi-level graph neural (HMGN) network that combines the CNN and graph neural network to capture both local and non-local image features at multiple scales. With the proposed patch graph attention module, the HMGN network can capture image features over a large receptive field, resulting in more accurate segmentation of cardiac structures. Experiments on two public datasets show the proposed method obtains improved segmentation performance over the state-of-the-art methods. |
|
3776.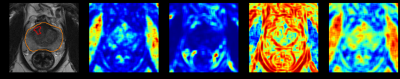 |
17 |
Deep Learning-based Prostate Lesion Segmentation and
Classification Using Haralick Texture Maps on MR images
Dang Bich Thuy Le1,
Ram Narayanan1,
Meredith Sadinski1,
Aleksandar Nacev1,
and Srirama Venkataraman1
1Research and Design, Promaxo Inc, Oakland, CA, United States Keywords: Radiomics, Cancer, Prostate By quantifying pixel relationships from frequencies of local signal intensity spatial variations, Haralick texture features have shown promise for prostate cancer detection. In this study, axial, T2-weighted MR images combined with extracted Haralick texture feature maps were used in a deep learning framework to identify lesion locations and predict Gleason Grade. Results demonstrate potential of Haralick texture features to segment and classify prostate lesions with AUC/sensitivity/specificity of 0.87/0.923/0.776 on patient-level evaluation. |
|
3777.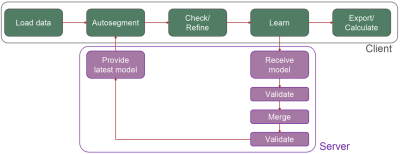 |
18 |
Lifelong collaborative learning improves the performance of
complex muscle MR image segmentation tasks
Francesco Santini1,2,
Jakob Wasserthal2,
Abramo Agosti3,
Xeni Deligianni1,2,
Kevin R Keene4,
Hermien E Kan5,
Stefan Sommer6,7,8,
Christoph Stuprich9,
Fengdan Wang10,
Claudia Weidensteiner1,11,
Giulia Manco12,
Valentina Mazzoli13,
Arjun Desai14,
and Anna Pichiecchio12,15
1Basel Muscle MRI, Department of Biomedical Engineering, University of Basel, Basel, Switzerland, 2Research Coordination Team, Department of Radiology, University Hospital Basel, Basel, Switzerland, 3Department of Mathematics, University of Pavia, Pavia, Italy, 4Department of Neurology, Leiden University Medical Center, Leiden, Netherlands, 5C.J. Gorter MRI Centre, Department of Radiology, Leiden University Medical Center, Leiden, Netherlands, 6Siemens Healthineers International AG, Zurich, Switzerland, 7Swiss Center for Musculoskeletal Imaging (SCMI), Balgrist Campus, Zurich, Switzerland, 8Advanced Clinical Imaging Technology (ACIT), Siemens Healthineers International AG, Lausanne, Switzerland, 9University Hospital Erlangen, Erlangen, Germany, 10Peking Union Medical College, Beijing, China, 11Radiological Physics, Department of Radiology, University Hospital Basel, Basel, Switzerland, 12Advanced Imaging and Radiomics Center, IRCCS Mondino Foundation, Pavia, Italy, 13Department of Radiology, Stanford University, Stanford, CA, United States, 14Departments of Electrical Engineering & Radiology, Stanford University, Stanford, CA, United States, 15Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy Keywords: Software Tools, Machine Learning/Artificial Intelligence An open-source, federated-learning-based segmentation software termed Dafne (Deep Anatomical Federated Network) is presented. This software continuously adapts the deep learning models used for the segmentation (currently for the muscles of the leg and thigh) based on the input of the users, who are in multiple institutions. This software was validated through data usage statistics of more than 50 users and through a retrospective study on 38 datasets of patients with suspected myositis, showing that the continuous learning approach is able to improve and generalize the performance of the original models. |
|
3778.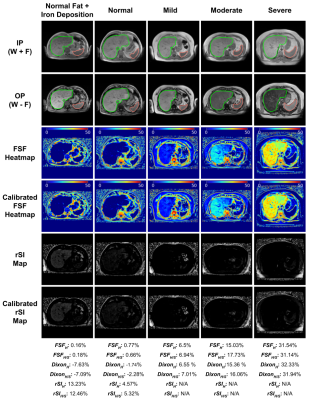 |
19 |
Deep Learning Based 3D Whole Liver and Spleen Segmentation for
Quantitatively Reproducible Liver Fat and Iron Deposition
Grading
Ahmed Gouda1,
Saqib Basar1,
Yosef Chodakiewitz2,
Rajpaul Attariwala1,
Sean London2,
and Sam Hashemi1
1Voxelwise Imaging Technology Inc., Vancouver, BC, Canada, 2Prenuvo, Vancouver, BC, Canada Keywords: Liver, Fat, Hepatic Steatosis Quantification, Iron Deposition Detection, Dixon, FSF, Dual-echo MRI Over the years, Magnetic Resonance Imaging (MRI) has become the optimal noninvasive method to quantify liver steatosis and to detect hepatic iron deposition. The conventional manual sampling technique of liver fat quantification at multiple regions is complex, inefficient and a time-consuming process. In addition, it may produce varying results for the heterogeneous fat deposition, which is prone to radiologist’s subjectivity. In this research, we propose a fully automated artificial intelligence (AI) based method for hepatic steatosis quantification and hepatic iron deposition detection using whole liver and spleen volume segmentation. |
|
3779.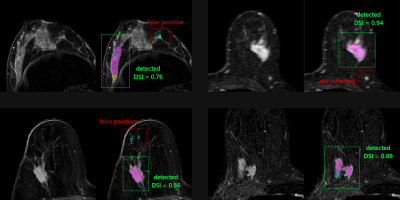 |
20 |
Automatic enhancing objects detection and segmentation in breast
DCE-MRI
Adam DESCARPENTRIES1,
Florence FERET2,
and Julien ROUYER1
1Research and Innovation Department, Olea Medical, La Ciotat, France, 2Clinical solution department, Olea Medical, La Ciotat, France Keywords: Breast, Cancer, CAD The aim of our work is to detect any enhancing object of interest for reporting purposes. A deep learning approach combined with a multi-constructor and multi-centric database enabled to initiate the development of a versatile tool in line with clinical real life. The detection problem was addressed using a two-stage three-dimensional cascaded U-Net architecture. A total of 610 single-breast images were used for the model development. Results present interesting score in term of Dice similarity index (0.83) which agree well with the recent literature. Discussion section focuses on the potential benefit in the use of a recently reported loss function. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.