Digital Poster
Software Tools
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
2391.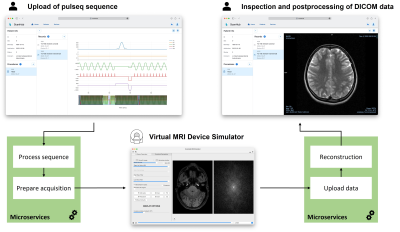 |
41 | ScanHub: Open-Source Platform for MR Scanner Control, Acquisitions and Postprocessing
David Schote1,2, Johannes Behrens2, Lukas Winter1, Christoph Kolbitsch1, and Christoph Dinh2
1Physikalisch-Technische Bundesanstalt (PTB), Braunschweig and Berlin, Germany, 2Brain-Link UG (haftungsbeschränkt), Landau i.d. Pfalz, Germany Keywords: Software Tools, Software Tools ScanHub (https://github.com/brain-link/scanhub-ui) is an open, generic solution for cloud-based medical data acquisition and processing. Functionalities are subdivided into microservices supporting use cases in the clinical as well as in the research context. The platform capabilities are demonstrated with an exemplary MRI workflow. As a proof of principle MR data acquisition was simulated with an open-source Bloch solver. A reconstruction of the simulated raw data is provided by a microservice. The whole acquisition process is controlled via a web-based UI; from deploying a pulse sequence to the organization and visualization of reconstructed and DICOMized results. |
|
2392.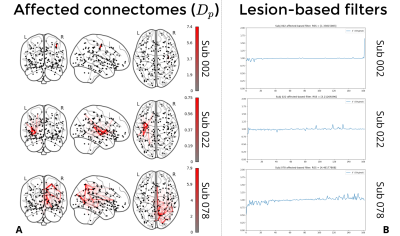 |
42 | NiGSP: Graph Signal Processing on multimodal MRI data.
Stefano Moia1,2, Julia Brügger1,3, Philip Egger1,3, Giorgia Giulia Evangelista1,3, Friedhelm Cristoph Hummel1,3,4, Maria Giulia Preti1,2, and Dimitri Van De Ville1,2
1Neuro-X Institute, Ecole polytechnique fédérale de Lausanne, Geneva, Switzerland, 2Department of Radiology and Medical Informatics (DRIM), Faculty of Medicine, University of Geneva, Geneva, Switzerland, 3Neuro-X Institute, EPFL Valais, Clinique Romande de Réadaptation, Sion, Switzerland, 4Department of Clinical Neurosciences, Geneva University Hospital (HUG), Geneva, Switzerland Keywords: Software Tools, Brain Connectivity NiGSP is a python-based toolbox aimed at facilitating the adoption of graph signal processing with an emphasis on multimodal brain imaging data. We present its standard workflow, that allows basic filtering operations and metrics computations, and we introduce a novel application to cerebral stroke consisting in the creation of a subject-specific anatomical lesion-based filter to be applied on functional MRI timeseries. |
|
2393. |
43 | CMRsim - A Python package for MRI simulations incorporating complex organ motion and flow
Jonathan Weine1, Charles McGrath1, and Sebastian Kozerke1
1University and ETH Zurich, Zurich, Switzerland Keywords: Software Tools, Simulations, Cardiovascular We present CMRsim, a new Python package for MR simulations efficiently incorporating complex organ motion and flow. The MR signal can be calculated using both Bloch simulations as well as analytical signal models. The package leverages TensorFlow2 for GPU acceleration and thereby facilitates fast simulations while also providing a compilation-free framework for prototyping and community contributions. Significant effort has been put on maintainability and software engineering best practices. The API reference as well as information on installation and how to get started is publicly available at: https://people.ee.ethz.ch/~jweine/cmrsim/latest/index.html Keywords: Open-source, Simulation, Cardiovascular, Motion, Reproducible science, Digital twin |
|
2394. |
44 | Open-source tools for multi-center multi-platform 0.55T relaxometry studies
Bilal Tasdelen1, Rajiv Ramasawmy2, Kathryn E Keenan3, Adrienne Campbell-Washburn2, and Krishna S Nayak1
1Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 2Cardiovascular Branch, Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, United States, 3NIST: National Institute of Standards and Technology, Boulder, CO, United States Keywords: Software Tools, Software Tools, open-source We demonstrate quantitative T1 and T2 mapping tools, built from open-source components Pulseq and qMRLab, suitable for multi-center multi-platform studies. The end-to-end solution includes acquisition, reconstruction, ROI analysis, and repeatability testing. The tools are demonstrated in the context of 0.55T relaxometry at two different sites and three scanners with two different hardware and software specifications. Relaxometry results are validated against results derived from vendor-provided sequences. |
|
2395. |
45 | Virtual Scanner Games
Gehua Tong1, Rishi Ananth2, Jason Stockmann3, Vlad Negnevitsky4, Benjamin Menküc5, Akbar Alipour6, John Thomas Vaughan, Jr.1,7, Sairam Geethanath7,8, Gaurav Verma9, and Gaurav Verma9
1Biomedical Engineering, Columbia University, New York, NY, United States, 2College of Arts and Sciences, University of Washington, Seattle, WA, United States, 3A.A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 4Oxford Ionics Ltd, Oxford, United Kingdom, 5University of Applied Sciences and Arts Dortmund, Dortmund, Germany, 6BioMedical Engineering and Imaging Institute, Dept. of Diagnostic, Molecular and Interventional Radiology, Icahn School of Medicine at Mt. Sinai, New York, NY, United States, 7Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States, 8Accessible MR Laboratory, BioMedical Engineering and Imaging Institute, Dept. of Diagnostic, Molecular and Interventional Radiology, Icahn School of Medicine at Mt. Sinai, New York, NY, United States, 9Radiology, Icahn School of Medicine at Mount Sinai, New York, NY, United States Keywords: Software Tools, Simulations, Education Access to MRI in low-resource regions is often limited by the lack of local expertise required to sustain long-term MR operation. To help develop interest and understanding in a wider audience from a younger age, we developed Virtual Scanner Games. This web application can be distributed by USBs to run locally and provides eight educational interactive games to introduce MR fundamentals to students at the high school level or higher. |
|
2396.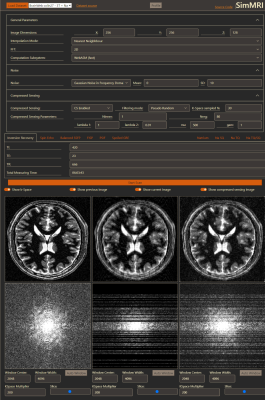 |
46 | SimMRI – A web-based MR Image Simulator for easy accessible MRI teaching
Christian Tönnes1, Christian Licht1, Lothar R. Schad1, and Frank G. Zöllner1
1Chair for Computer Assisted Clinical Medicine, MIiSM, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany Keywords: Software Tools, Simulations, Education SimMRI is a Web-Based MRI simulator developed for teaching students. It allows the simulation of multiple sequences for 1H and 23Na imaging on different brain datasets. The software enables users to customize parameters for each sequence, add noise, or change the voxel count for every dimension of the image. Additionally, the software provides compressed sensing reconstruction with three k-space subsampling schemes. The computation is solely performed on the client and therefore well suited for large student groups. The code, written in JavaScript, HTML, CSS, and c compiled to WebASM, is open source and supports easy inclusion of new sequences. |
|
2397.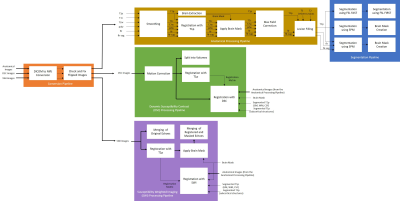 |
47 | Magnetic Resonance Image Processing and Analysis Platform (MRI-PAP): A Windows-based Automated Pipeline for Multiple Sclerosis
Alia Khaled1,2, Ahmed Bayoumi1, Joseph Thomas1, Refaat Gabr3, Khader Hasan3, Jerry Wolinsky1, and John Lincoln1
1Department of Neurology, University of Texas Health Sciences Center, Houston, TX, United States, 2Department of Cancer Systems Imaging, University of Texas MD Anderson Cancer Center, Houston, TX, United States, 3Department of Diagnostic and Interventional Imaging, University of Texas Health Sciences Center, Houston, TX, United States Keywords: Software Tools, Data Processing, Pipeline Neurologists and researchers rely on several software tools for neuroimaging data analysis. Limitations in these tools often necessitate the use of more than one program, spanning different coding languages and operating systems, which requires time, effort, and programming skills. MRI-PAP is a collection of pipelines with an easy-to-use graphical user interface (GUI) designed for MRI processing and analysis of multiple sclerosis patient images. These automated pipelines accept DICOM images as input and take them through a series of processing steps. MRI-PAP pipelines save time by accessing different packages from a single GUI and can be used without programming skills. |
|
2398. |
48 | CMRSeq - A Python package for intuitive sequence design
Jonathan Weine1, Charles McGrath1, and Sebastian Kozerke1
1University and ETH Zurich, Zurich, Switzerland Keywords: Software Tools, Pulse Sequence Design We present CMRseq, a new python package for MR sequence definitions. The package builds upon successful concepts of previous frameworks, while leveraging Python functionality to increase usability and maintainability. CMRseq features physical unit checks, adheres to coding style conventions and includes unit-test coverage and continuous integration infrastructure for documentation. Import and export functionality to facilitate compatibility with popular formats is also included. The API reference as well as information on installation and how to get started is publicly available at: https://people.ee.ethz.ch/~jweine/cmrseq/latest/index.html Keywords: Open source, reproducible science, sequence design, software tools, visualization |
|
2399.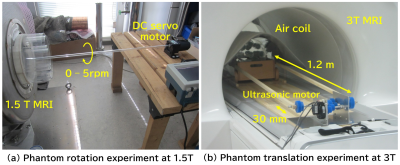 |
49 | MRI simulation of moving objects based on the Lagrange description of Bloch equations
Katsumi Kose1, Ryoichi Kose1, Daiki Tamada2, and Utaroh Motosugi3
1MRIsimulations Inc., Tokyo, Japan, 2University of Yamanashi, Chuo, Japan, 3Kofu Kyoritsu Hospital, Kofu, Japan Keywords: Software Tools, Motion Correction, motion simulation To investigate the possibility and limitation of MRI simulations based on the Lagrange description, MRI experiments and simulations of moving objects were performed. As a result, we were able to obtain simulated MR images that almost reproduced the experimental results within practical computation time. However, it was found that the T2 coherence effect should be reduced for moving objects. It was also found that more precise simulation is necessary to reproduce the detailed motion artifacts. |
|
2400.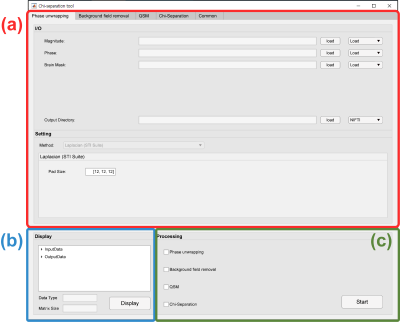 |
50 | A toolbox of chi-separation for magnetic susceptibility source separation
Jun-Hyeok Lee1, Hyeong-Geol Shin2,3, Minjun Kim2, Jongho Lee2, and Se-Hong Oh1
1Biomedical Engineering, Hankuk University of Foreign Studies, Yongin, Korea, Republic of, 2Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of, 3Johns Hopkins University School of Medicine & Kennedy Krieger Institute, Baltimore, MD, United States Keywords: Software Tools, Quantitative Susceptibility mapping, Chi-separation The susceptibility maps generated by QSM and $$$\chi$$$-separation show magnetic susceptibility distributions, which have been proposed as important biomarkers for brain disorders. However, susceptibility mapping requires a complicated multi-step procedure that is difficult for inexperienced researchers. There is a need to design a convenient application that can easily conduct susceptibility mapping. In this work, we developed a MATLAB based GUI software, called the ‘$$$\chi$$$-separation toolbox’ that generates high-quality susceptibility maps in just a few clicks. The GUI of our toolbox is intuitive and user-friendly that it helps researchers to conduct QSM and $$$\chi$$$-separation processing without difficulty. |
|
2401.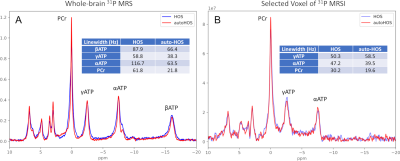 |
51 | Automated High Order Shim for Neuroimaging Studies
Jia Xu1, Baolian Yang2, Douglas Kelley2, and Vincent A. Magnotta1,3,4
1Radiology, University of Iowa, Iowa City, IA, United States, 2GE Healthcare, Waukesha, WI, United States, 3Psychiatry, University of Iowa, Iowa City, IA, United States, 4Biomedical Engineering, University of Iowa, Iowa City, IA, United States Keywords: Software Tools, Shims In this work, we proposed an automated High Order Shim procedure for neuroimaging studies. The proposed shimming procedure is fully automated and hence eliminates variability between operators. The procedure performs automated real-time brain extraction to define the region of interest (ROI) of the field map to be used in the shimming algorithm. Automated High Order Shim has fewer image distortions and narrower spectral linewidths than linear shimming and manual high-order shimming, suggesting its superior performance in correcting B0 field homogeneity. The shimming performance was assessed by acquiring EPI-based images and MR spectroscopy at both 3T and 7T field strengths. |
|
2402.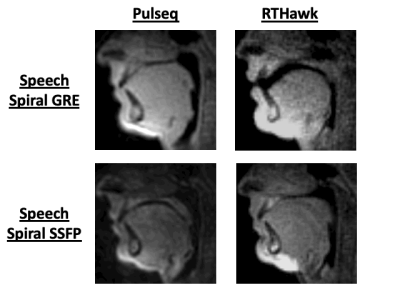 |
52 | Open-source dynamic MRI workflow for reproducible research
Prakash Kumar1, Bilal Tasdelen1, and Krishna S Nayak1
1Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States Keywords: Software Tools, Data Acquisition This work presents an open-source workflow for dynamic MR acquisition and reconstruction. The entire workflow consists of vendor-agnostic tools to allow for immediate sharing of data, pulse sequence, and reconstruction code across sites. This work is an adaptation of Open-Source MR Imaging and Reconstruction Workflow to dynamic imaging (Veldmann et. al, MRM 88(6):2395-2407). Using the FIRE interface, we demonstrate a Gadgetron loader of acquisition metadata including k-space trajectories and present a pipeline to convert pulse sequences developed through HeartVista’s SpinBench into Pulseq for easy sharing of sequences. |
|
2403.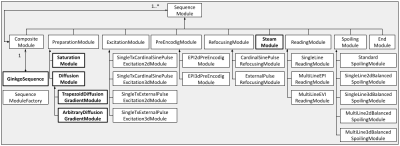 |
53 | A diffusion-weighted MRI pulse sequence development toolbox in the open source GinkgoSequence framework
Anaïs Artiges1, Éléa Granier1, Ivy Uszynski1, Franck Mauconduit1, Philippe Ciuciu2, and Cyril Poupon1
1BAOBAB, NeuroSpin, Paris-Saclay University, CNRS, CEA, Gif-sur-Yvette, France, 2Mind, NeuroSpin, Paris-Saclay University, INRIA, CEA, Gif-sur-Yvette, France Keywords: Software Tools, Pulse Sequence Design Developing MRI pulse sequences demands to access proprietary sequences, to open up parts of the code provided by the manufacturer, and to deal with intellectual property issues. To address these limitations, we have provided the community with GinkgoSequence, a modular and open-source MRI pulse sequence development framework for Siemens devices. In this work, we present the addition of a new diffusion-weighted sequences development toolbox in the existing GinkgoSequence framework. It includes trapezoidal and free-waveform diffusion gradients, fat saturation, echo-planar reading, partial Fourier and GRAPPA accelerations, and stimulated echo acquisition mode (STEAM). To assess its quality, it has been tested in-vivo. |
|
2404.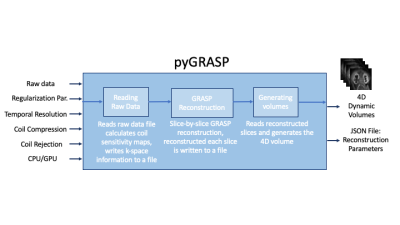 |
54 | PyGRASP: A standalone python image reconstruction library for DCE-MRI acquired with radial sampling
Cemre Ariyurek1, Aziz Koçanaoğulları1, Can Taylan Sari1, Serge Vasylechko1, Onur Afacan1, and Sila Kurugol1
1Quantitative Intelligent Imaging Lab (QUIN), Department of Radiology, Boston Children's Hospital and Harvard Medical School, Boston, MA, United States Keywords: Software Tools, DSC & DCE Perfusion Dynamic contrast enhanced MRI (DCE-MRI) is capable of quantitative assessment of renal function and evaluation of the detailed anatomy of the urinary tract and renal arteries in patients. To reconstruct the dynamic volumes in DCE-MRI, Golden-angle RAdial Sparse Parallel (GRASP) reconstruction algorithm is commonly used. In this software, we have developed an efficient open-source, purely Python-based, standalone GRASP reconstruction library called pyGRASP that allows researchers to access the source code for development, facilitating flexible deployment with readable code and no compilation, and easy utilization without requirement of a steep learning curve. |
|
2405.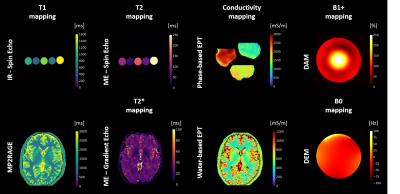 |
55 | qMRpy: a cross-platform, extensible and easy-to-use open-source Python toolbox for efficient estimation of quantitative MR properties
Matteo Cencini1, Marta Lancione1, Luca Peretti1, and Michela Tosetti1
1IRCCS Stella Maris, Pisa, Italy Keywords: Software Tools, Software Tools Quantitative MRI methods improve tissue characterization and allow for better longitudinal assessment, but need efficient and reliable fitting routines. Currently available qMR frameworks, while covering a wide range of applications, suffer either from a relatively low efficiency or require non-trivial compilation steps. Here, we propose an open-source framework for qMR quantification which maintains a good computational efficiency without sacrificing the readability and customizability of the code. |
|
2406.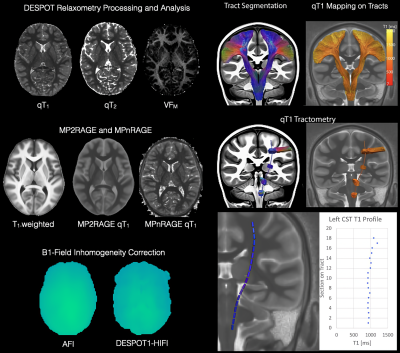 |
56 | QMRI-neuropipe: A flexible software framework for the analysis of quantitative MRI data
Douglas C Dean III1,2,3, Nagesh Adluru3, and Jose Guerrero3
1Pediatrics, University of Wisconsin–Madison, Madison, WI, United States, 2Medical Physics, University of Wisconsin–Madison, Madison, WI, United States, 3Waisman Center, University of Wisconsin–Madison, Madison, WI, United States Keywords: Software Tools, Data Processing Quantitative MRI provides a unique opportunity to characterize the underlying tissue and establish measurements that canserve as biomarkers. However, many of these methods require specialized workflows and and tools which limit their broader adoption. Here, we present QMRI-neuropipe, an open-source, flexible framework that provides a wide selection of methods, algorithms, and tools for processing and analyzing multiple qMRI datatypes. QMRI-neuropipe supports the Brain Imaging Datset Standard (BIDS) and currently supports processing of diffusion and relaxometry datasets. Future developments will aim to incorporate alternative quantitative MRI methods (e.g. MT, QSM, etc.) into the QMRI-neuropipe framework. |
|
2407.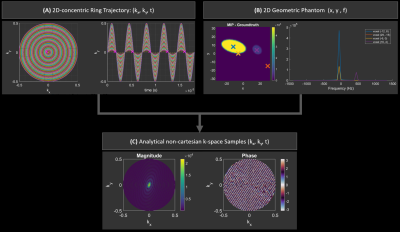 |
57 | Simulation tool for non-Fourier MRSI reconstruction
Carina Graf1 and Christopher T Rodgers2
1Wolfson Brain Imaging Centre, Department of Clinical Neuroscience, University of Cambridge, Cambridge, United Kingdom, 2University of Cambridge, Cambridge, United Kingdom Keywords: Software Tools, Spectroscopy, non-cartesian MRSI With the wider availability of ultra-high field MR systems, metabolic imaging using MRSI is a fast-developing area of research. Frequently used sequences use non-uniformly sampled trajectories to achieve high-acceleration factors e.g. concentric-ring trajectories (CRT). In this work, we demonstrate the from-scratch implementation of a simulation and reconstruction tool for CRT-MRSI using (1) a regridding, (2) an iterative L2 linear solver as well as (3) applying the non-uniform FFT implementation from the BART-toolbox to simulated non-cartesian MRSI data with a known Fourier-transform pair. The exact sampled data permits the evaluation of the impact of different reconstruction implementations. |
|
2408.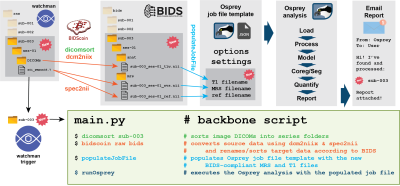 |
58 | Continuous automated MRS data analysis workflow
Aaron T. Gudmundson1,2, Helge J. Zöllner1,2, Christopher W. Davies-Jenkins1,2, Erik G. Lee3,4, Timothy J. Hendrickson3,4, Richard A. E. Edden1,2, and Georg Oeltzschner1,2
1The Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Kennedy Krieger Institute, F. M. Kirby Research Center for Functional Brain Imaging, Baltimore, MD, United States, 3Masonic Institute for the Developing Brain, University of Minnesota, Minneapolis, MN, United States, 4Informatics Institute, University of Minnesota, Minneapolis, MN, United States Keywords: Software Tools, Spectroscopy, Automation Large-scale application of magnetic resonance spectroscopy (MRS) is limited by demanding data processing and reliance on expert knowledge. We have developed an open-source continuous automated analysis workflow for MRS data, substantially reducing the effort for manual data analysis and quality control. This workflow is of particular interest for application-oriented non-expert users and large-scale multi-center multi-vendor MRS studies, and should substantially protect against loss of data from acquisition protocol errors. |
|
2409.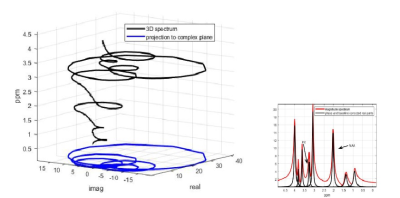 |
59 | Novel 3D Post-Processing for 1H Magnetic Resonance Spectroscopy: Fast and Accurate Metabolite Ratios
Dale H. Mugler1
1Neuroscience, Medical University of South Carolina, Charleston, SC, United States Keywords: Software Tools, Spectroscopy, Data Analysis, Metabolism Brain tumor metabolic maps are one application of non-invasive MR Spectroscopy (MRS), although there are many other areas of patient treatment and surgery planning that would benefit from improved MRS analysis. A fast, accurate new method for MRS is used here to estimate the ratio of Choline to NAA from simulated spectra modeled on those near a brain tumor, using simple formulas for determining the FID amplitudes that relate to metabolite intensities and concentrations. No approximate numerical integration is required. The time of computation of 0.052 seconds per voxel speeds the construction of a metabolic 3D brain map. |
|
2410.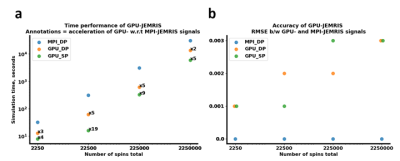 |
60 | GPU-accelerated JEMRIS for extensive MRI simulations.
Aizada Nurdinova1,2, Stefan Ruschke2, Jonathan Stelter2, Michael Gestrich3, and Dimitrios Karampinos2
1Biomedical Physics, Stanford University, Stanford, CA, United States, 2Department of Diagnostic and Interventional Radiology, School of Medicine, Technical University of Munich, Munich, Germany, 3Altair Engineering GmbH, Boeblingen, Germany Keywords: Simulations, Software Tools Bloch simulations is a powerful tool in MRI research and education as it allows predicting the signal evolutions in magnetic resonance experiments for various applications. However, Bloch simulations require solving ordinary differential equations for the whole spin ensemble and can thus become infeasible for realistic scenarios with large total number of spins. Therefore, the present work investigates the potential of a GPU-based implementation for accelerated Bloch simulations as an extension to the open source Juelich Extensible MRI Simulator (JEMRIS) in comparison to the existing MPI CPU implementation. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.