Digital Poster
ML/AI for Segmentation & Registration I
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
4342.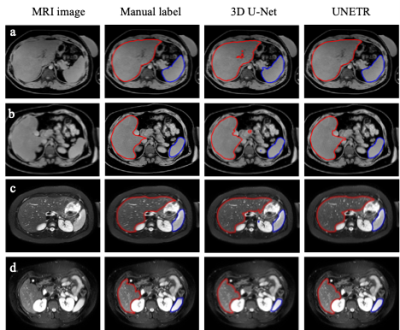 |
81 |
Simultaneous Liver and Spleen Segmentation using U-Net
Transformer Model on T1-weighted and T2-weighted MRI Data
Huixian Zhang1,
Redha Ali1,
Hailong Li1,
Wen Pan1,
Scott B. Reeder2,
David T. Harris2,
William R. Masch3,
Anum Alsam3,
Krishna P. Shanbhogue4,
Nehal A. Parikh1,
Jonathan R. Dillman1,
and Lili He1
1Cincinnati Children's Hospital, Cincinnati, OH, United States, 2University of Wisconsin-Madison, Madison, WI, United States, 3Michigan Medicine, University of Michigan, Ann Arbor, MI, United States, 4NYU Langone Health, New York, NY, United States Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence To develop an AI system for precise and fully automated, simultaneous segmentation of the liver and spleen from T1-weighted and T2-weighted MRI data. Our study compares the performance of the U-Net Transformer (UNETR) and the standard 3D U-Net model for simultaneous liver and spleen segmentation using MRI images from pediatric and adult patients from multiple institutions. Our work demonstrates that the UNETR shows a statistical improvement over 3D U-Net in both liver and spleen segmentations. |
|
4343.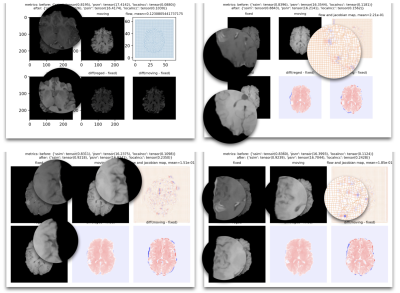 |
82 |
A Deep Learning-based Multi-Contrast MRI Registration Model with
a Realistic Flow Field and Reduced Over-Smoothing Effect
Yiheng Li1 and
Ryan Chamberlain1
1Subtle Medical Inc., Menlo Park, CA, United States Keywords: Machine Learning/Artificial Intelligence, Data Processing, Registration To optimize DL-based medical registration models to produce a more realistic flow field and reduce the over-smoothing effect of deformable registration while keeping the generalizability of the multi-contrast registration model to work on all anatomical structures and contrast of MRI, we proposed an algorithm that adopts the image synthetic framework from SynthMorph and optimized with cycle-consistent loss. By experimenting with the jacobian loss, bidirectional loss, and cycle-consistent loss, we managed to further optimize the results of registered images. The evaluation of two MRI image datasets, the BraTS dataset, and the LSpine dataset, demonstrated the increased SSIM, PSNR, and LocalNCC. |
|
4344.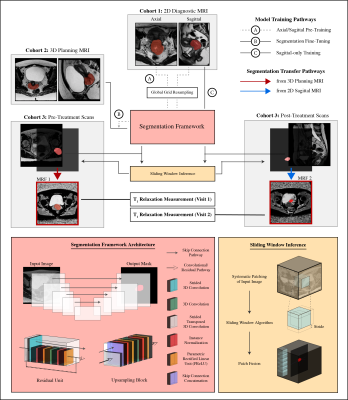 |
83 |
Spatial-Adaptive Deep Learning Model and Magnetic Resonance
Fingerprinting for Segmentation and Quantitative Evaluation of
Cervical Cancer
Reza Kalantar1,
Jessica Mary Winfield1,2,
Mihaela Rata1,
Gigin Lin3,
Susan Lalondrelle1,4,
Christina Messiou1,2,
Matthew David Blackledge1,
and Dow-Mu Koh1,2
1Radiotherapy and Imaging, The Institute of Cancer Research, London, United Kingdom, 2Radiology, The Royal Marsden Hospital, London, United Kingdom, 3Medical Imaging and Intervention, Chang Gung Memorial Hospital at Linkou and Chang Gung University, Taoyuan, Taiwan, 4Gynaecological Unit, The Royal Marsden Hospital, London, United Kingdom Keywords: Machine Learning/Artificial Intelligence, Cancer Quantitative magnetic resonance imaging (qMRI) can provide additional information for diagnosis and response assessment, but adoption of multi-parametric qMRI techniques has been hindered by long acquisition times and labor-intensive processing steps. Magnetic resonance fingerprinting (MRF) provides quantitative maps in a single acquisition but MRF deployment in clinical studies still requires manual delineation of volumes of interest. A spatial-adaptive deep learning framework was developed to segment cervical cancer on MRI and quantify T1 relaxation times of the tumor pre- and post-radiotherapy treatment. Our results suggest that automated segmentation models may be promising tools for quantitative tumor evaluation and treatment response assessment. |
|
4345.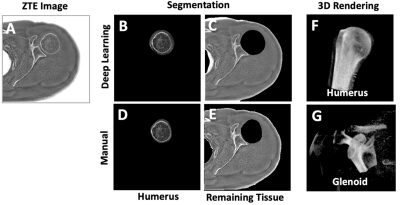 |
84 |
ZTE segmentation of glenohumeral bone structure using deep
learning
Michael Carl1,
Kaustaub Lall2,
Armin Jamshidi2,
Eric Chang3,4,
Sheronda Statum3,4,
Anja Brau1,
Christine B Chung3,4,
Maggie Fung1,
and Won C. Bae3,4
1General Electric Healthcare, Menlo Park, CA, United States, 2Electrical and Computer Engineering, University of California, San Diego, La Jolla, CA, United States, 3Radiology, University of California, San Diego, La Jolla, CA, United States, 4Radiology, VA San Diego Healthcare System, San Diego, CA, United States Keywords: Machine Learning/Artificial Intelligence, Segmentation, ZTE, Deep learning Evaluation of 3D bone morphology of the glenohumeral joint is necessary for pre-surgical planning. Zero echo time (ZTE) MRI provides excellent bone contrast, and we developed a deep learning model to perform automated segmentation of major bones (i.e., humerus and others) from ZTE to aid evaluation. Axial ZTE images of normal shoulders (n=31) acquired at 3T were annotated for training with a 2D U-Net, and the trained model was validated with testing data (n=10 normal shoulder, n=6 symptomatic). Testing accuracy was around 80 to 90% (Dice score) for either cohort, except for a few failed cases with very low scores. |
|
4346. |
85 |
Novel Uunsupervised Segmentation of Bone Marrow Edema-Like
Lesions using Bayesian Conditional Generative Adversarial
Networks
Andrew Seohwan Yu1,2,3,
Sibaji Gaj1,2,
William Holden1,2,
Richard Lartey1,2,
Jeehun Kim1,2,4,
Carl Winalski1,2,5,
Naveen Subhas1,2,5,
and Xiaojuan Li1,2,5
1Program of Advanced Musculoskeletal Imaging (PAMI), Cleveland Clinic, Cleveland, OH, United States, 2Department of Biomedical Engineering, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, United States, 3Cleveland Clinic, Department of Computer and Data Sciences, Case Western Reserve University, Cleveland, OH, United States, 4Department of Electrical, Computer, and Systems Engineering, Case Western Reserve University, Cleveland, OH, United States, 5Department of Diagnostic Radiology, Imaging Institute, Cleveland Clinic, Cleveland, OH, United States Keywords: Machine Learning/Artificial Intelligence, Segmentation Quantitative assessment of the bone marrow edema-like lesions and their association with osteoarthritis requires a consistent and unbiased segmentation method, which is difficult to obtain in the presence of human annotators. This study proposes an unsupervised approach using Bayesian deep learning and conditional generative adversarial networks that detects and segments anomalies without human intervention. The full pipeline has a lesion-wide sensitivity of 0.86 on unseen scans. This approach is expected to be generalizable to other lesions and/or modalities. |
|
4347.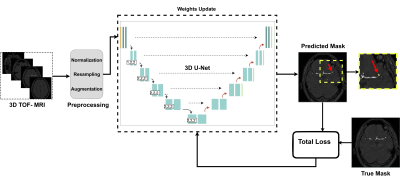 |
86 |
3D nnU-Net with Multi Loss Ensembles for Automated Segmentation
of Intracranial Aneurysm
Maysam Orouskhani1,
Shaojun Xia2,
Mahmud Mossa-Basha1,
and Chengcheng Zhu1
1Department of Radiology, University of Washington, Seattle, WA, United States, 2Peking University Cancer Hospitals & Institution, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Compound Loss function, Deep Neural Networks, nnU-Net In the segmentation of intracranial aneurysm, deep neural networks are equipped with modified loss functions to penalize the training weights for aneurysm false predictions and conduct unbiased learning. In this paper, we used a new compound loss function to capture the different aspects of embedding as well as diverse features. The proposed loss was given to a 3D full resolution nnU-Net to segment imbalanced TOF-MRA images from ADAM dataset. The proposed loss outperformed commonly used losses in terms of Dice, Sensitivity, and Precision. |
|
4348.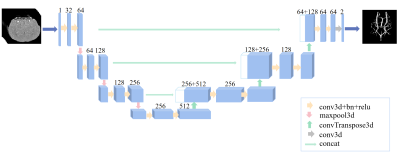 |
87 |
Cerebrovascular segmentation of mouse TOF-MRA using 3D U-Net
Yue Wu1,2,
Jinyuan Zhang3,4,
Zhixin Li3,4,
Yishuang Yang3,4,
Yan Zhuo3,4,
Xudong Zhao2,3,
and Zihao Zhang2,3
1AHU-IAI AI Joint Laboratory, Anhui University, Hefei, China, 2Institute of Artificial Intelligence, Hefei Comprehensive National Science Center, Hefei, China, 3State Key Laboratory of Brain and Cognitive Science, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China, 4University of Chinese Academy of Sciences, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Vessels Time-of-flight MR angiography (TOF-MRA) allows noninvasive visualization of the mouse intracranial vessels. However, there is no available tool for vessel segmentation in mouse 3D TOF-MRA. In this study, we developed an automated vessel segmentation method based on a convolutional neural network and multiscale vessel enhancement filtering algorithm. The method is potentially useful in the preclinical studies of cerebrovascular diseases. |
|
4349.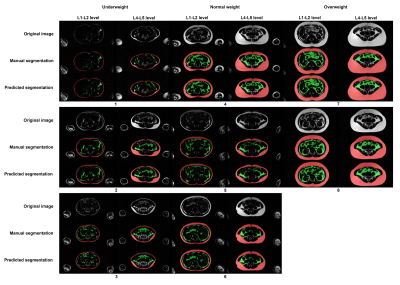 |
88 |
Automated deep learning based segmentation of abdominal adipose
tissue on whole-body MRI in a population-based study of
adolescents
Tong Wu1,
Santiago Estrada2,
Renza Gils1,
Ruisheng Su1,
Vincent Jaddoe3,
Edwin Oei1,
and Stefan Klein1
1Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, Netherlands, 2Image Analysis, German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany, 3Department of Pediatrics, Erasmus MC, Rotterdam, Netherlands Keywords: Machine Learning/Artificial Intelligence, Segmentation, Dixon MRI This study is embedded in the Generation R Study, a population-based prospective cohort study in the Netherlands. Whole-body Dixon MRI scans were performed at age 13 years. A previous neural network (CDFNet) has been published that was trained on adults. We aimed to retrain this network on our MRIs in the Generation R Study. The obtained segmentations are in strong agreement with expert-generated manual segmentations and can therefore greatly reduce the manual workload. Therefore, for accurate abdominal fat quantification, segmentation of both subcutaneous and visceral adipose tissue, on Dixon MRIs of adolescents is feasible with a retrained convolutional neural network. |
|
4350.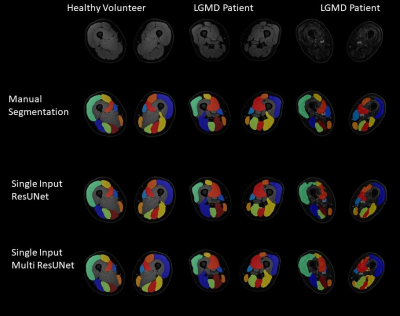 |
89 |
3D segmentation of MRI images of leg muscles using Convolutional
Neural Networks
Lara Schlaffke1,
Niklas Doliwa1,
Marius Markmann1,
Marlena Rohm1,2,
Robert Rehmann1,3,
and Martijn Froeling4
1Neurology, University Clinic Bergmannsheil Bochum gGmbH, Bochum, Germany, 2Heimer Institute for Muscle Research, University Clinic Bergmannsheil Bochum gGmbH, Bochum, Germany, 3Neurology, Klinikum Dortmund, University Witten-Herdecke, Dortmund, Germany, 4Radiology, UMC Utrecht, Utrecht, Netherlands Keywords: Machine Learning/Artificial Intelligence, Muscle, Segmentation Quantitative magnetic resonance imaging offers promising surrogate biomarkers for the early diagnosis and monitoring of pathological changes in leg muscles in patients with neuromuscular disorders. To use this in a clinical routine, automatic segmentation of muscles is needed. To investigate precision and accuracy of various Computational neuronal networks for automated segmentation of upper leg muscles, different loss function were used and quantitative outcome compared to the gold-standard of manual segmentation. The best segmentation results were achieved, with the Fokal Tversky loss function and a single input approach. |
|
4351.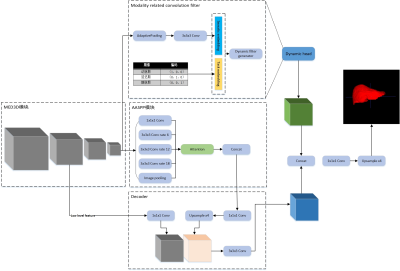 |
90 |
A deep learning-based liver tumor segmentation algorithm in
enhanced multi–phase MRI images
Meng Dou1,2,
Ying Zhao 3,
Tao Lin3,
Yu Yao1,2,
and Ailian Liu3
1Chengdu institute of computer application, Chinese academy of sciences, Chengdu, China, 2University of Chinese Academy of Sciences, Beijing, China, 3Department of Radiology, First Affiliated Hospital of Dalian Medical University, Dalian, China Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence In this paper, we propose a deep learning-based liver tumor segmentation algorithm in enhanced multi-phase MRI images. The experimental results show that the proposed method can segment liver and tumor in enhanced multi-phase MRI images with a smaller resource occupation, and outperforms the comparison method. |
|
4352.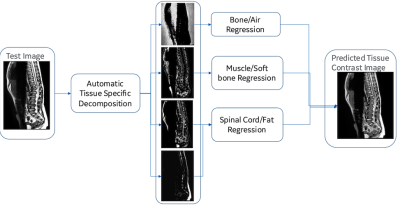 |
91 |
A Non-linear, Tissue-specific 3D Data Generative Framework for
Deep Learning Segmentation Performance Enhancement
Soumya Ghose1,
Chitresh Bhushan1,
Dattesh Shanbhag2,
Desmond Teck Beng Yeo3,
and Thomas K. Foo3
1AI and Computer Vision, GE Research, Niskayuna, NY, United States, 2GE Healthcare, Bangalore, India, 3Biology & Physics, GE Research, Niskayuna, NY, United States Keywords: Machine Learning/Artificial Intelligence, Segmentation, Vertebra segmentation, T2w, Deep Learning, Intensity Transformation Deep learning (DL) models have been successful in solving segmentation problems that involve large, balanced, and labeled datasets. However, in the medical imaging domain, it is rare to find manually annotated datasets that capture the entire spectrum of heterogeneity. Novel datasets with significantly different intensities than training datasets, may adversely affect DL model performance. In this work, we present a hybrid framework for tissue-specific, non-linear intensity transformation of pediatric T2w images similar to that of the adults training dataset and demonstrate an improved performance for vertebra segmentation of pediatric datasets without the need for DL network re-training/re-tuning. |
|
4353.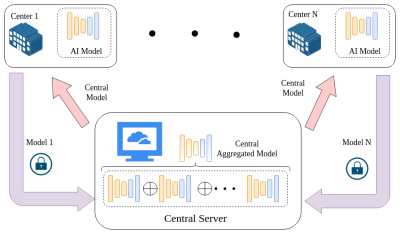 |
92 |
A novel federated learning framework for accurate and secure
multi-center MS lesion segmentation
Dongnan Liu1,2,
Mariano Cabezas2,
Dongang Wang2,3,
Zihao Tang1,2,
Geng Zhan2,3,
Kain Kyle2,3,
Linda Ly2,3,
James Yu2,3,
Chun-Chien Shieh2,3,
Ryan Sullivan4,
Fernando Calamante4,5,
Michael Barnett2,3,
Wanli Ouyang6,
Weidong Cai1,
and Chenyu Wang2,3
1the School of Computer Science, University of Sydney, Sydney, Australia, 2Brain and Mind Centre, University of Sydney, Sydney, Australia, 3Sydney Neuroimaging Analysis Centre, Sydney, Australia, 4the School of Biomedical Engineering, University of Sydney, Sydney, Australia, 5Sydney Imaging, University of Sydney, Sydney, Australia, 6the School of Electrical and Information Engineering, University of Sydney, Sydney, Australia Keywords: Machine Learning/Artificial Intelligence, Segmentation Multiple sclerosis (MS) is a neurodegenerative disease of the central nerve system (CNS), which has the potential to cause a neurological disability, particularly for young adults. Recently, deep learning-based techniques are important for MS diagnosis and treatment, since they can segment the lesions caused by MS automatically and accurately. However, their applicability to multi-center scenarios is limited, due to the privacy and security issues in data sharing. To tackle these limitations, a decentralized deep learning framework is designed in this work, which can bring accurate multi-center MS lesion segmentation performance without sharing the raw data. |
|
4354.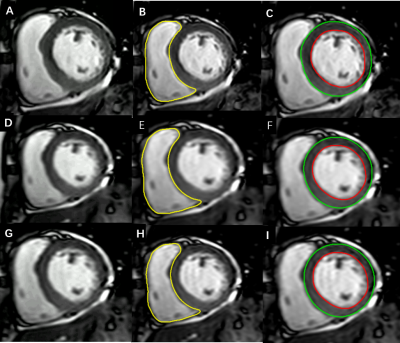 |
93 |
Accelerated cardiac cine imaging using deep learning: impact on
left ventricular function and AI segmentation model
Wenyun Liu1,
Chinting Wong2,
Cheng Li3,
Di Yu1,
Yi Zhu4,
Ke Jiang4,
and Huimao Zhang1
1Radiology, The First Hospital of Jilin University, Changchun, China, 2Nuclear Medicine, The First Hospital of Jilin University, Changchun, China, 3Cardiovascular Center, The First Hospital of Jilin University, Changchun, China, 4Philips Healthcare, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Myocardium, Cardiomyopathy,Data Analysis Cardiac MRI is considered reference standard for the noninvasive assessment of ventricular volumes and function. However, long multibreath-hold acquisition time can prove difficult in patients and lead to poor image quality. In this study, we investigated the use of a deep learning-based reconstruction algorithm, named Compressed SENSE Artificial Intelligence(CS-AI), to accelerate two-dimensional cine bSSFP for cardiac MRI. The purpose of this study was to compare the image quality and performance of a CS-AI-based cine sequence between reference and accelerated methods: SENSE, Compressed-SENSE, and CS-AI, and then to investigate the impact of images reconstructed by deep learning on AI segmentation model. |
|
4355.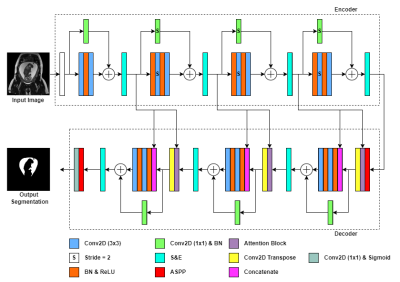 |
94 |
Amniotic Fluid Segmentation using Convolutional Neural Networks
Alejo Costanzo1,2,
Birgit Ertl-Wagner3,4,
and Dafna Sussman1,5,6
1Department of Electrical, Computer and Biomedical Engineering, Toronto Metropolitan University, Toronto, ON, Canada, 2Institute for Biomedical Engineering, Science and Technology, Toronto Metropolitan University and St. Michael’s Hospital, Toronto, ON, Canada, 3Division of Neuroradiology, The Hospital for Sick Children, Toronto, ON, Canada, 4Department of Medical Imaging, Faculty of Medicine, University of Toronto, Toronto, ON, Canada, 5Institute for Biomedical Engineering, Science and Technology, Toronto Metropolitan University and St. Michael’s Hospital, Toronto, ON, Canada, 6Department of Obstetrics and Gynecology, Faculty of Medicine, University of Toronto, Toronto, ON, Canada Keywords: Machine Learning/Artificial Intelligence, Segmentation, Convolutional Neural Network Amniotic Fluid Volume (AFV) is an important fetal biomarker when diagnosing certain fetal abnormalities. We aim to implement a novel Convolutional Neural Network (CNN) model for amniotic fluid (AF) segmentation which can facilitate clinical AFV evaluation. The model, called AFNet was trained and tested on a radiologist–validated AF dataset. AFNet improves upon ResUNet++ through the efficient feature mapping in the attention block, and transpose convolutions in the decoder. Experimental results show that our AFNet model achieved a 93.38% mean Intersection over Union (mIoU) on our dataset. We further demonstrate that AFNet outperforms state-of-the-art models while maintaining a low model size. |
|
4356.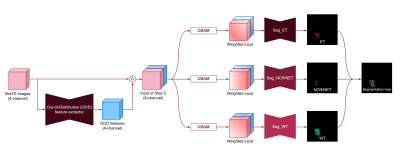 |
95 |
Brain tumor segmentation using out-of-distribution feature from
normal MR images
Namho Jeong1,
Beomgu Kang1,
and Hyunwook Park1
1Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of Keywords: Machine Learning/Artificial Intelligence, Tumor Abundant data of healthy subjects is available and thus it would be helpful if normal data can boost the brain tumor segmentation. We proposed the out-of-distribution (OOD) feature based on the learned distribution of normal data to discriminate abnormal tumors. Once the neural network was trained to synthesize other contrast MR images with normal data, it failed to properly synthesize the tumor tissues. This OOD characteristic was used for the generation of new feature that can help the tumor segmentation. It was verified that the OOD features contributed to an improvement of segmentation through experiments. |
|
4357.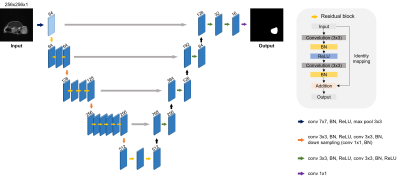 |
96 |
Breast Tumor Segmentation using U-Net with ResNet34
Yunkyoung Jun1,
Jiwoo Jeong1,
Seokha Jin1,
Noehyun Myung1,
Jimin Lee2,
and Hyungjoon Cho1
1BME, UNIST, Ulsan, Korea, Republic of, 2Nuclear Engineering, UNIST, Ulsan, Korea, Republic of Keywords: Machine Learning/Artificial Intelligence, Animals The proposed research implemented an automatic tumor segmentation application on an orthotopic breast tumor model. This application can segment the tumors accurately and monitor tumor growth and the therapeutic effect of Doxorubicin for treatment. Also, the outputs from the application can reconstruct into 3D rendering and offer the visualization of shape and volume. As a result, the application can be applied to orthotopic breast tumor model research. |
|
4358.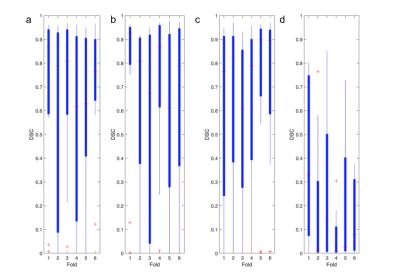 |
97 |
Comparison of Combinations of MTR, FLAIR, and T1WCE MRI Images
for Automatic Segmentation of Brain Metastases
Ananya Anand1,
Daniel Nussey2,
and Dafna Sussman3
1Toronto Metropolitan University, Fremont, CA, United States, 2Toronto Metropolitan University, Toronto, ON, Canada, 3Electrical, Computer, and Biomedical Engineering, Toronto Metropolitan University, Toronto, ON, Canada Keywords: Machine Learning/Artificial Intelligence, Magnetization transfer, Multi-Modal This study presents an evaluation of utilizing MTR and FLAIR as compared to T1wCE and FLAIR images for automatic segmentation of brain metastases, due to the unknown consequences posed by accumulated contrast enhancement. Numerous combinations of FLAIR, T1wCE, and MTR MRI images were tested on three top-performing segmentation models (MS-FCN, U-NET, and SegresNet) to evaluate the feasibility of using MTR and FLAIR images for tumor segmentation. Overall, the U-Net produced the best similarity scores of 0.4±0.15 using MTR and FLAIR images. A future study will test the utility of 3D MTR images for tumor segmentation using convolutional and full-connected models. |
|
4359.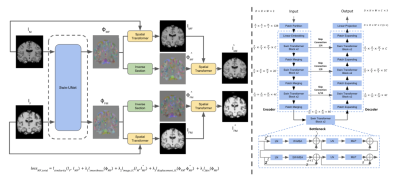 |
98 |
Cycle Inverse Consistent Deformable Medical Image Registration
with Transfomer
Chenghao Zhang1,
Mikhail Morgan1,
Yanting Yang1,
Ye Tian1,
and Jia Guo1
1Columbia University, New York, NY, United States Keywords: Machine Learning/Artificial Intelligence, Brain Deformable image registration is a crucial part of the medical image process that seeks to estimate an optimal spatial transformation to align two images. Traditional methods neglect the inverse-consistent property and topology preservation of the transformation, which can lead to errors in registration results. To address these issues, we propose the cycle inverse consistent transformer-based deformable medical image registration model. We adopt Swin-UNet to achieve higher registration performance and comprehensively consider the inverse consistency loss function to guarantee a more accurate registration. Our pipeline can be trained to create study-specific templates of images for diagnostic and/or educational purposes. |
|
4360. |
99 |
Deep learning Based Automatic Segmentation of Brain Magnetic
Resonance angiography Images
Liying An1,
Fanyang Meng1,
Kaixin Yu2,
Lei Zhang1,
and Huimao Zhang1
1Radiology, The First Hospital of Jilin University, Changchun, China, 2Shukun (Beijing) Technology Co., Ltd, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Vessels Accurate cerebrovascular segmentation plays an important role in clinical diagnosis and the related research. Magnetic Resonance angiography (MRA) postprocessing manually recognized by technologists is extremely labor intensive and error prone. However, automatic segmentation of brain vessels remains challenging because of the variable vessel shape and high complex of vessel geometry. We propose an artificial intelligence brain vessel segmentation system supported by an optimized physiological anatomical-based 3D convolutional neural network that can automatically achieve brain MRA vessel segmentaion in healthcare services. The overall segmentation accuracy of the independent testing dataset is 0.931. |
|
4361.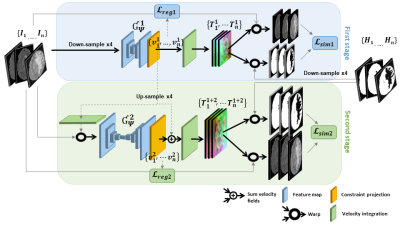 |
100 |
Deep learning-based groupwise registration for longitudinal MRI
analysis in glioma
Claudia Chinea Hammecher1,2,
Karin van Garderen1,3,
Marion Smits1,3,
Pieter Wesseling4,5,
Bart Westerman6,
Pim French7,
Mathilde Kouwenhoven8,
Roel Verhaak9,10,
Frans Vos1,2,
Esther Bron1,
and Bo Li1
1Department of Radiology & Nuclear Medicine, Erasmus MC, Rotterdam, Netherlands, 2Department of Imaging Physics, Delft University of Technology, Delft, Netherlands, 3Medical Delta, Delft, Netherlands, 4Department of Pathology, Amsterdam UMC / VU Medical Center, Amsterdam, Netherlands, 5Laboratory for Childhood Cancer Pathology, Princess Máxima Center for Pediatric Oncology, Utrecht, Netherlands, 6Department of Neurosurgery, Amsterdam UMC / VU Medical Center, Amsterdam, Netherlands, 7Department of Neurology, Erasmus MC Cancer Institute, Rotterdam, Netherlands, 8Department of Neurology, Amsterdam UMC / VU Medical Center, Amsterdam, Netherlands, 9The Jackson Laboratory for Genomic Medicine, Farmington, CT, United States, 10Department of Neurosurgery, Amsterdam UMC/VUmc, Amsterdam, Netherlands Keywords: Machine Learning/Artificial Intelligence, Data Analysis, Image Registration Glioma growth may be quantified with longitudinal image registration. However, the large mass-effects and tissue changes across images suposse an added challenge. Here, we propose a longitudinal, learning-based and groupwise registration method for the accurate and unbiased registration of glioma MRI. We evaluate on a dataset from the Glioma Longitudinal AnalySiS consortium and compare to classical registration methods. We achieve comparable Dice coeffients, with more detailed registrations, while significantly reducing the runtime to under a minute. The proposed methods may serve as an alternative to classical toolboxes, to provide further insight into glioma growth . |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.