Digital Poster
ML/AI for General Image Analysis & Post-Processing I
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
4732.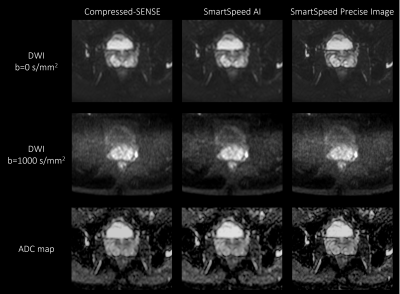 |
141 |
Deep learning-based image quality and spatial resolution
improvement in Turbo Spin Echo Diffusion Weighted Imaging for
prostate
Jihun Kwon1,
Kohei Yuda2,
Masami Yoneyama1,
Yasutomo Katsumata3,
and Marc Van Cauteren3
1Philips Japan, Tokyo, Japan, 2Tokyo Metropolitan Police Hospital, Nakano, Japan, 3Philips Healthcare, Best, Netherlands Keywords: Prostate, Diffusion/other diffusion imaging techniques Diffusion-weighted imaging (DWI) plays an important role in assessing the significance of prostate cancer. DWI with Turbo Spin Echo readout (TSE-DWI) is robust to image distortion but suffers from low signal to noise ratio. In this study, we investigated the use of prototype AI-based reconstruction technique (SmartSpeed Precise Image) to improve the image quality of TSE-DWI images. The image quality was compared between conventional Compressed-SENSE (C-SENSE), SmartSpeed AI, and SmartSpeed Precise Image. Volunteer data demonstrated a significant improvement of sharpness in both b=0 and 1000 s/mm2 images as well as ADC map, compared with C-SENSE and SmartSpeed AI reconstructions. |
|
4733.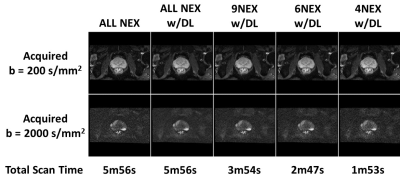 |
142 |
Quantitative Analysis and Scan Time Impact of DL Recon Applied
to Single-Shot Diffusion of the Prostate
Eugene Milshteyn1,
Arnaud Guidon1,
and Mukesh G. Harisinghani2
1GE Healthcare, Boston, MA, United States, 2Massachusetts General Hospital, Boston, MA, United States Keywords: Prostate, Diffusion/other diffusion imaging techniques Increasing the speed of multiparametric prostate MRI (mpMRI) is highly desirable. However, usual tradeoffs between signal-to-noise (SNR) and scan time must be considered and impact on quantitative metrics must be analyzed. One recently proposed approach applied a commercialized deep learning reconstruction (DL Recon) to prostate T2-weighted imaging, leveraging the capabilities of the DL algorithm to achieve a robust, high-quality T2-weighted acquisition in half the time. As such, this work focuses on evaluating the DL Recon on diffusion weighted imaging, which shows promise to cut acquisition time by ~70% and therefore benefit mpMRI. |
|
4734.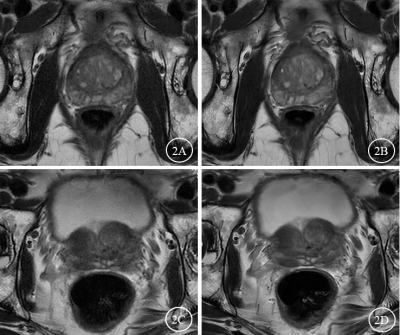 |
143 |
Deep learning-based high-resolution T2-weighted imaging elevated
clarity of prostatic calcification and diagnostic efficacy
Zan Ke1,
Liang Li1,
Zhi Wen1,
Weiyin Vivian Liu2,
and Yunfei Zha1
1Radiology, Renmin Hospital of Wuhan University, Wuhan, China, 2GE Healthcare, MR Research China, Beijing, China Keywords: Prostate, Prostate Prostatic calcification is common in benign prostatic hyperplasia (BPH) and usually asymptomatic. Our study showed the prostate T2WIDL images have higher subjective rating scores, clearer lesion contrast and improved detection rate of prostatic calcification, higher SNR and CNR. In addition, T2WIDL more clearly and sharply displayed prostate capsule, lesion contrast, prostate calcification and anatomical details Therefore, AIR™ Recon DL based T2WI (T2WIDL) quality in prostate MRI offer higher overall image quality and elevated a younger radiologist’s diagnostic performance on prostate calcification. |
|
4735.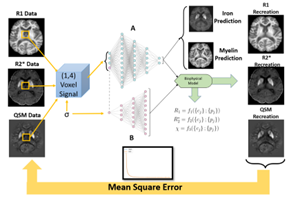 |
144 |
Conventional χ-separation compared to self calibrated method (BIOPHYSICSS-DL)
with histological validation
Ilyes Benslimane1,
Günther Grabner2,
Simon Hametner3,
Thomas Jochmann1,4,
Robert Zivadinov1,5,
and Ferdinand Schweser1
1Department of Neurology, Buffalo Neuroimaging Analysis Center, Buffalo, NY, United States, 2Department of Medical Engineering, Carinthia University of Applied Sciences, Klagenfurt, Austria, 3Department of Neuropathology and Neurochemistry, Medical University of Vienna, Vienna, Austria, 4Department of Computer Science and Automation, Technische Universität Ilmenau, Ilmenau, Germany, 5Department of Computer Science and Automation, Center for Biomedical Imaging, Clinical and Translational Science Institute at the University at Buffalo, Buffalo, NY, United States Keywords: Machine Learning/Artificial Intelligence, Brain The χ-separation method determines para- and diamagnetic susceptibility tissue compartments correlating to iron and myelin in the brain respectively. The method presupposes subject invariant relaxometry coefficients and compartments disregarding the changes in those parameters in disease or postmortem cases. We implement a biophysically informed autoencoder network developed for single subject use (BIOPHYSICSS-DL) to determine underlying biophysical model coefficients from individual datasets. We expand the current model with different combinations of relaxometry and susceptibility data to produce a self-calibrated χ separation method finding the network comparable to standard methods for iron and predicts myelin distribution more closely to ground truth histology. |
|
4736.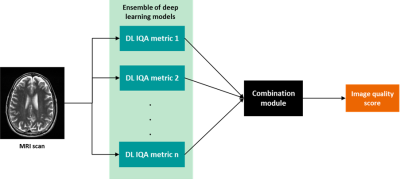 |
145 |
Motion artifact assessment for magnetic resonance imaging using
a learned combination of deep learning model predictions.
Vanya Saksena1,2,
Silvia Arroyo-Camejo2,
Julian Hossbach1,2,
Rainer Schneider2,
and Andreas Maier1
1Pattern Recognition Lab, Friedrich-Alexander-University Erlangen-Nuremberg, Erlangen, Germany, 2Siemens Healthineers, Erlangen, Germany Keywords: Machine Learning/Artificial Intelligence, Artifacts, Image quality Magnetic resonance imaging (MRI) is a powerful imaging modality, but susceptible to various image quality problems. Today, technicians conduct image quality assurance (IQA) during scan-time as a manual, time-consuming, and subjective process. We propose a method towards adaptable automated IQA of MR images without the need of a large, annotated image database for training. Our method implements a machine learning-based module that uses multiple predictions from an ensemble of deep learning models trained with image quality metrics. The sensitivity of this method to detect image quality problems is adaptable to clinical requirements of the end user. |
|
4737.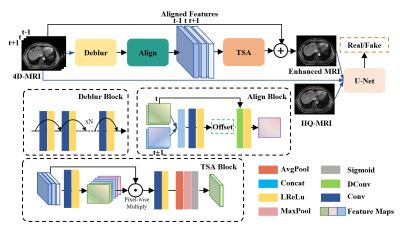 |
146 |
Implicit Temporal-compensated Adversarial Network for 4D-MRI
Enhancement
Yinghui Wang1,
Tian Li1,
Haonan Xiao1,
and Jing Cai1
1Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, Hong Kong Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, 4D-MRI\Enhancement\Temporal-compensation In this study, we proposed and evaluated a deep learning technique for refining four-dimensional magnetic resonance imaging (4D-MRI) in the post-processing stage. More specifically, we designed an implicit temporal-compensated adversarial network (ITAN) based on the intrinsic property of 4D-MRI to improve image quality with neighboring phases. It can overcome its inherent challenges of data deficiency, misalignment between training pairs, and complex texture details. The qualitative and quantitative results demonstrated that the proposed model can suppress the noise and artifacts in 4D-MR images, recover the missing details and perform better than a state-of-the-art method. |
|
4738.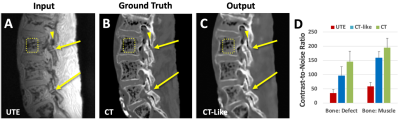 |
147 |
Deep learning tools to aid the evaluation of isthmic
spondylolysis
Vadim Malis1,
Suraj Achar2,
Dosik Hwang3,
and Won C. Bae1,4
1Radiology, University of California, San Diego, La Jolla, CA, United States, 2Family Medicine, University of California, San Diego, La Jolla, CA, United States, 3Electrical and Electronics Engineering, Yonsei University, Seoul, Korea, Republic of, 4VA San Diego Healthcare System, San Diego, CA, United States Keywords: Machine Learning/Artificial Intelligence, Bone, UTE, Deep Learning, Image Regression, Saliency Map Isthmic spondylolysis results in fracture of pars interarticularis of the lumbar spine in young athletes. UTE MRI provide good bone contrast, although CT is still the gold standard. To take UTE MRI further, we developed supervised deep-learning tools to generate CT-like images and saliency maps of fracture probability from UTE MRI and CT, using ex vivo preparation of cadaveric spines. The results demonstrate feasibility of CT-like images to provide easier interpretability for bone fractures, due to improved image contrast and CNR, and the saliency maps to aid in quick detection of pars fracture by providing visual cues to the reader. |
|
4739.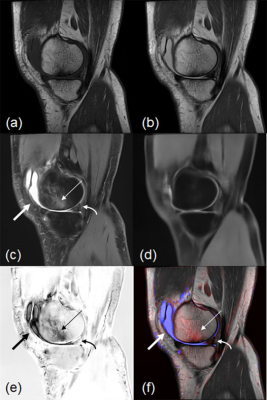 |
148 |
A Fat-suppression, Image-subtraction Method Using Deep Learning
for the Detection of Knee Abnormalities on MRI
Tsutomu Inaoka1,
Akihiko Wada2,
Tomoya Nakatsuka1,
Masayuki Sugeta1,
Akinori Yamamoto1,
Hisanori Tomobe1,
Ryousuke Sakai1,
Hiroyuki Nakazawa1,
Masaru Sonoda3,
Rumiko Ishikawa1,
Shusuke Kasuya1,
and Hitoshi Terada1
1Radiology, Toho University Sakura Medical Center, Sakura, Japan, 2Radiology, Juntendo University, Tokyo, Japan, 3Radiology, Seirei Sakura Citizen Hospital, Sakura, Japan Keywords: Machine Learning/Artificial Intelligence, Joints This fat-suppression subtraction-image method using a DL model with 2D CNNs may be useful for the detection and classification of abnormalities on knee MRI. |
|
4740.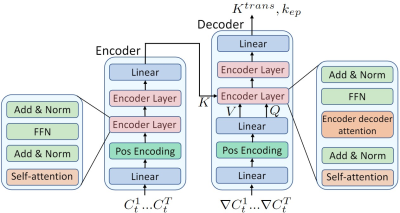 |
149 |
Accelerating DCE-MRI Analysis for Prostate Cancer Diagnosis with
Deep Neural Networks
Kai Zhao1,
Haoxin Zheng1,
and Kyunghyun Sung1
1Department of Radiological Sciences, University of California, Los Angeles, Los Angeles, CA, United States Keywords: Machine Learning/Artificial Intelligence, Data Analysis, dynamic contrast enhanced A deep learning based DCE-MRI analysis method was proposed with a dedicated neural network architecture and data generation framework. The proposed method does not need DCE-MRI data acquisition or annotation for training. Compared to conventional non-linear least square (NLLS) fitting methods, the proposed method significantly reduced the average processing time from hours to few minutes while preserved the estimation quality. |
|
4741.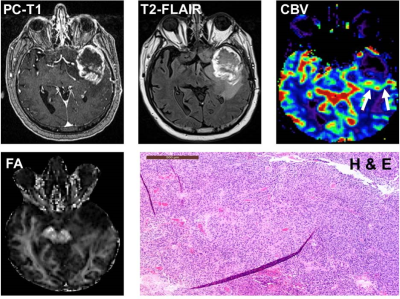 |
150 |
Distinction of True-Progression from Pseudo-progression in
Glioblastomas using ML Model based on Quantitative mpMRI and
Molecular Signatures
Virendra Kumar Yadav1,
Suyash Mohan2,
Sumeet Kumar Agarwal3,4,
Laiz Laura de Godoy2,
Sumei Wang2,
MacLean P. Nasrallah5,
Donald M. O’Rourke6,
Stephen Bagley7,
Harish Poptani8,
Sanjeev Chawla2,
and Anup Kumar Singh1,4,9
1Centre for Biomedical Engineering, Indian Institute of Technology, Delhi, India, 2Departments of Radiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States, 3Department of Electrical Engineering, Indian Institute of Technology, Delhi, India, 4Yardi School of Artificial Intelligence, Indian Institute of Technology, Delhi, India, 5Clinical Pathology and Laboratory, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States, 6Neurosurgery, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States, 7Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States, 8Department of Molecular and Clinical Cancer Medicine, University of Liverpool, United Kingdom, Liverpool, United Kingdom, 9Department of Biomedical Engineering, All India Institute of Medical Sciences, New Delhi, India Keywords: Machine Learning/Artificial Intelligence, Brain Glioblastoma patients (n=93) exhibiting enhancing lesions within 6 months after completion of standard therapy underwent anatomical imaging, diffusion and perfusion MRI. The median values of parameters (MD, FA, CL, CP, CS and rCBV) were computed from the enhancing regions. O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation status was available from 75 patients. Subsequently, these patients were classified as TP (n=55) or PsP (n=20). The data were randomly split into training and testing sets. The best model for differentiating TP from PsP was obtained using quadratic SVM classifier with a training accuracy of 90.9%, cross-validation accuracy of 85.5% and testing accuracy of 85%. |
|
4742.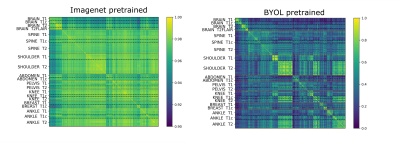 |
151 |
Anatomy and Image Contrast Metadata Verification using
Self-supervised Pretraining
Ben A Duffy1 and
Ryan Chamberlain1
1Subtle Medical Inc., Menlo Park, CA, United States Keywords: Machine Learning/Artificial Intelligence, Data Processing Automated metadata verification is essential for data quality checking. This study evaluates the extent to which self-supervised pretraining can improve performance on the anatomy and image contrast metadata verification tasks. On a small but diverse dataset, pretraining coupled with supervised finetuning, outperforms training from an ImageNet initialization, suggesting improved near out-of-distribution performance. On a larger brain-only dataset, training a linear classifier on the self-supervised pretrained network embeddings outperforms the corresponding ImageNet pretraining or random initialization on the image contrast prediction task. Cross-checking predictions against the DICOM metadata was an effective method for detecting artifacts and other quality issues. |
|
4743.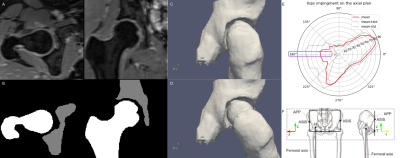 |
152 |
A neural network to estimate the hip center of rotation for a
fully-automated range of motion analysis in femoroacetabular
impingement.
Eros Montin1,2,
Daniele Panozzo3,
and Riccardo Lattanzi1,2,4
1Center for Advanced Imaging Innovation and Research (CAI2R) Department of Radiology, Radiology Department, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States, 2Bernard and Irene Schwartz Center for Biomedical Imaging, Department of Radiology, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States, 3New York University, New York, New York, USA, BROOKLYN, NY, United States, 4Vilcek Institute of Graduate Biomedical Sciences, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States Keywords: Machine Learning/Artificial Intelligence, Joints We evaluated three neural network architectures for the automatic identification of the center of the femur head on 3D water-only Dixon MRI. We trained using a mixture of real and augmented data. The mean error of the best-performing network was three-time lower compared to a manual annotation and on the order of 1 voxel. We combined the network to create the first fully automated pipeline to assess the hip range of motion from 3D MR. |
|
4744.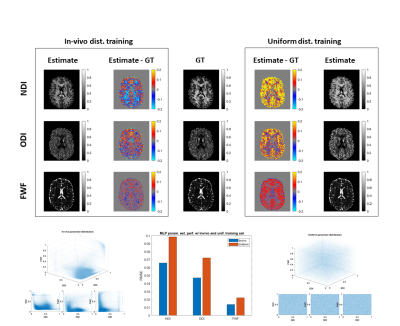 |
153 |
Can machine learning resolve model degeneracy in tissue
microstructure estimation?
Michele Guerreri1,2,
Sean Epstein1,
Hojjat Azadbakht2,
and Hui Zhang1
1Computer Science & Centre for Medical Image Computing, University College London, London, United Kingdom, 2AINOSTICS Ltd., Manchester, United Kingdom Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence This work investigates the impact of model degeneracy on machine learning-based tissue microstructure estimation. While there have been several empirical reports suggesting machine learning can not resolve model degeneracy, the impact of model degeneracy is poorly understood. Here we show how model degeneracy can be categorised into three types with varying degrees of impact on machine learning-based microstructure estimation. Our finding is important for designing optimal training data distribution. |
|
4745. |
154 |
A Deep Learning Nomogram Based on Gd-EOB-DTPA MRI for Predicting
Early Recurrence in Hepatocellular Carcinoma after Hepatectomy
Meng Yan1,
Xiao Zhang2,
Bin Zhang1,
Zhendong Qi3,
Xiaoyun Liang2,
Feng Huang2,
Shuixing Zhang1,
Xinming Li3,
Shutong Wang4,
and Xianyue Quan3
1Department of Radiology, The First Affiliated Hospital of Jinan University, Guangzhou, China, 2Neusoft Medical Systems Co., Ltd, Shanghai, China, 3Department of Radiology, Zhujiang Hospital of Southern Medical University, Guangzhou, China, 4Department of Liver Surgery, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China Keywords: Machine Learning/Artificial Intelligence, Cancer Prognostic risk assessment after hepatectomy for patients with hepatocellular carcinoma (HCC) remains difficult. Previous studies have shown that Gd-EOB-DTPA MRI is sensitive and accurate for HCC detection, but studies in predicting early recurrence after hepatectomy based on deep learning (DL) are still lacking. This study investigated the performance of a Gd-EOB-DTPA MRI-based DL approach, and then evaluated the DL nomogram incorporating deep features and significant clinical indicators. DL nomogram outperformed the clinical nomogram (validation AUC: 0.909 vs. 0.715). The proposed DL nomogram could provide a noninvasive and comprehensive tool for predicting early recurrence of HCC after curative resection. |
|
4746.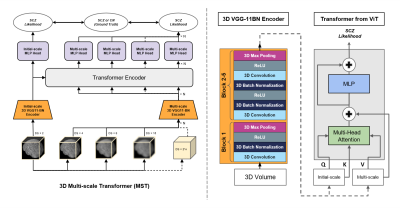 |
155 |
Improving Across-Dataset Schizophrenia Classification with
Structural Brain MRI Using Multi-scale Transformer
Ye Tian1,
Junhao Zhang2,
Vish Mitnala Rao1,
and Jia Guo3
1Biomedical Engineering, Columbia University, New York, NY, United States, 2BME, Columbia University, New York, NY, United States, 3Department of Psychiatry, Columbia University, New York, NY, United States Keywords: Machine Learning/Artificial Intelligence, Brain, Deep Learning Schizophrenia is a neurological disorder that requires accurate and rapid detection for earlier intervention. Previous explorations in artificial intelligence showed overwhelming performance using deep learning in schizophrenia classification, though the generalization remained a challenge. We propose our 3D Multi-scale Transformer (MST) using T1W structural MRI data to detect schizophrenia. By synthesizing reconstructed images at different scales, the transformer-based architecture improves robustness to generalize in unseen data. The proposed method reaches the same-level performance of AUROC to the benchmark mark model in schizophrenia identification, and performs better in all leave-one-site-out generality tests. |
|
4747.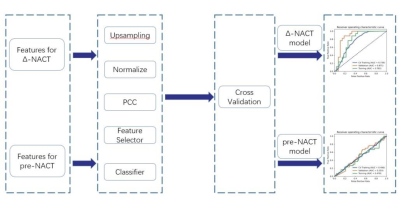 |
156 |
Using machine learning to evaluate values of six diffusion
models to predict the efficacy of neoadjuvant chemotherapy for
esophageal cancer
Long Cui1,
Bingmei Bai2,
Chenglong Wang1,
Yang Song3,
Shengyong Li1,
Haijie Wang1,
Jinrong Qu2,
and Guang Yang1
1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 2Department of Radiology, Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, China, 3MR Scientific Marketing, Siemens Healthcare, Shanghai, China Keywords: Cancer, Tumor We assessed the performance of diffusion models for assessing response to neoadjuvant chemotherapy (NACT) using machine learning. Firstly, features were extracted from the region of interest on different parametric maps of different diffusion models for esophageal squamous cell carcinoma (ESCC) patients and changes of the parameters (Δ parameter) before and after NACT (pre-NACT and post-NACT) were calculated. Then different Δ-NACT models and pre-NACT models were built for using features from different diffusion models. The results demonstrated that diffusion models may be used to predict the efficacy of NACT in ESCC patients. |
|
4748.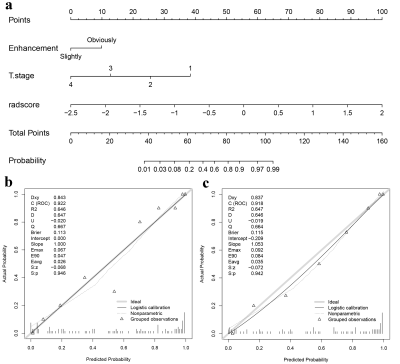 |
157 |
Multi-parametric MRI Radiomics Combined with Clinical Factors to
Predict the Response to Neoadjuvant Chemotherapy in
Nasopharyngeal Carcinoma
Zhuo Wang1,
Zhiqiang Chen2,
Xiaohua Chen1,
Shaoru Zhang1,
Yunshu Zhou1,
Ruodi Zhang1,
Shili Liu1,
Yuhui Xiong3,
and Bing chen4
1Department of Clinical Medicine of Ningxia Medical University, Yinchuan, China, 2Department of Radiology, the First Hospital Affiliated to Hainan Medical College, Haikou, China, 3GE Healthcare, Beijing, China, 4General Hospital of Ningxia Medical University, Yinchuan, China Keywords: Cancer, Head & Neck/ENT, multi-parametric MRI-based radiomics This study aims to evaluate the predictive performance of the nomogram integrating multi-parametric MRI-based radiomics and clinical characteristics in detecting therapeutic response to neoadjuvant chemotherapy (NAC) in nasopharyngeal carcinoma (NPC) patients. Least absolute shrinkage and selection operator (LASSO) regression was performed to select radiomics features. A nomogram was constructed using multivariable logistic regression. The receiver operating characteristic (ROC) curves, calibration, and decision curves were performed to assess the discriminative performance of the clinical, radiomics, and the combined models. The nomograms developed by integrating radiomics score (Rad-Score) with clinical factors outperformed the clinical model alone. |
|
4749.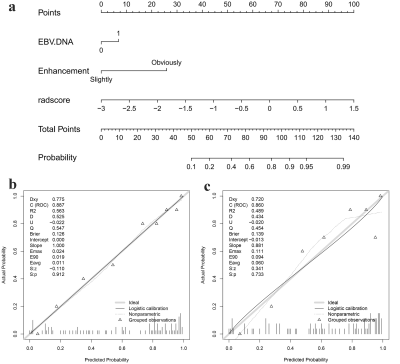 |
158 |
Multi-parametric MRI-based Radiomics integrated with Clinical
Features for Predicting the Ki-67 Labeling Index in
Nasopharyngeal Carcinoma
Zhuo Wang1,
Zhiqiang Chen2,
Xiaohua Chen1,
Shaoru Zhang1,
Shili Liu1,
Ruodi Zhang1,
Yunshu Zhou1,
Yuhui Xiong3,
and AiJun Wang4
1Department of Clinical Medicine of Ningxia Medical University, Yinchuan, China, 2the First Hospital Affiliated to Hainan Medical College, Haikou, China, 3GE Healthcare, Beijing, China, 4General Hospital of Ningxia Medical University, Yinchuan, China Keywords: Cancer, Head & Neck/ENT, multi-parametric MRI-based radiomics To investigate the value of a nomogram based on multi-parametric MRI-based radiomics combined with clinical imaging features in predicting the Ki-67 LI in nasopharyngeal carcinoma. Least absolute shrinkage and selection operator (LASSO) regression was performed to select radiomics features. A nomogram was established using multivariable logistic regression. The receiver operating characteristic (ROC) curves, calibration, and decision curves were performed to evaluate the predictive performance of the different models. The nomograms constructed by integrating radiomics score (Rad-Score) with clinical imaging factors outperformed the clinical or the radiomics models alone. |
|
4750.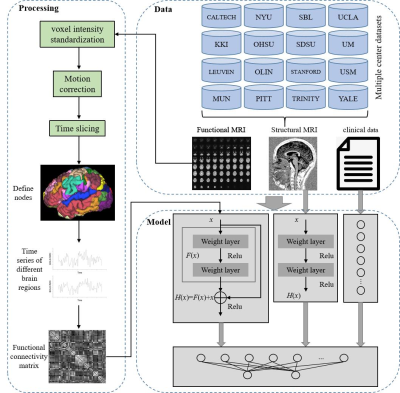 |
159 |
Multi-branch deep learning model integrated with multi-modal MRI
for differential diagnosis of autism spectrum disorders
Xuan Yu1 and
Meiyun Wang1,2
1Henan Provincial People’s Hospital & the People’s Hospital of Zhengzhou University, Zhengzhou, China, 2Laboratory of Brain Science and Brain-Like Intelligence Technology, Institute for Integrated Medical Science and Engineering, Henan Academy of Sciences, Zhengzhou, China Keywords: Brain Connectivity, Brain Connectivity This study proposes a multi-branch deep learning model fused with multi-modal MRI for the differential diagnosis of ASD. We solve the problem that traditional ASD classification algorithms based on static functional network connections ignore the time-varying characteristics of brain functional connections. Study the spatiotemporal characteristics of ASD brain imaging, and mine the information of functional connectivity between brain regions over time. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.