Digital Poster
ML/AI for Segmentation & Registration II
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
4511.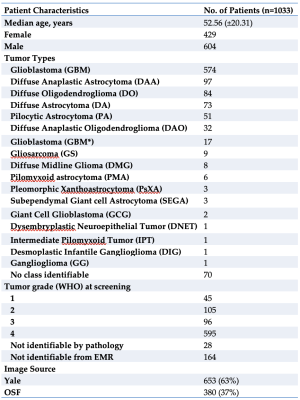 |
81 |
The Yale Glioma Dataset: Developing An Open Access, Annotated
MRI Database
Matthew L. Sala1,
Jan Lost2,
Niklas Tillmanns2,
Sara Merkaj3,
Marc von Reppert4,
Divya Ramakrishnan1,
Khaled Bousabarah5,
Anita Huttner6,
Sanjay Aneja7,
Arman Avesta7,
Antonio Omuro8,
and Mariam Aboian1
1Radiology and Biomedical Imaging, Yale School of Medicine, New Haven, CT, United States, 2University of Düsseldorf, Düsseldorf, Germany, 3University of Ulm, Ulm, Germany, 4Leipzig University, Leipzig, Germany, 5Visage Imaging, Düsseldorf, Germany, 6Pathology, Yale School of Medicine, New Haven, CT, United States, 7Therapeutic Radiology, Yale School of Medicine, New Haven, CT, United States, 8Neurology, Yale School of Medicine, New Haven, CT, United States Keywords: Tumors, Machine Learning/Artificial Intelligence, Glioma Recent development of Machine Learning (ML) tools for analysis of CNS tumors demonstrates great potential benefit to research and clinical practice but has been hindered by a lack of external validation. There is a critical need for open access to large individual hospital-based datasets with expert annotations. Here, we present the Yale Glioma Dataset, a database of 1,033 patients featuring annotated segmentations on FLAIR and T1 post-gadolinium, tumor grading and classification, and further clinical information. Open access of this database will support the development and validation of new AI algorithms for glioma detection and segmentation. |
|
4512.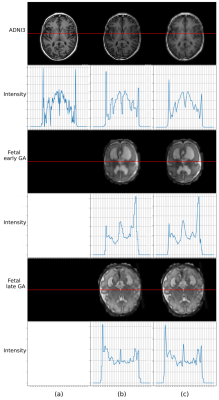 |
82 |
Efficient Fetal Brain Segmentation according to the Point Spread
Function of MRI
Yunzhi Xu1,
Jiaxin Li1,
Xue Feng2,
Kun Qing3,
Dan Wu1,
and Li Zhao1
1Zhejiang University, Hangzhou, China, 2Biomedical Engineering, University of Virginia, Virginia, VA, United States, 3Department of Radiation Oncology, City of Hope National Center, Los Angeles, CA, United States Keywords: Machine Learning/Artificial Intelligence, Fetus, Fetal Brain MRI,Point Spread Function High apparent resolution of fetal MRIs is provided by slice-to-volume reconstruction pipelines widely. However, the physical resolution of the fetal brain is lower than that. Therefore, we hypothesize that fetal brain segmentation can be performed based on downsampled fetal brain MRI according to its point spread function. In this work, 150 adult brain and 80 fetal brain MRIs were used to validate hypothesize. Using downsampled fetal data with factor of 4, a highly efficient segmentation model achieved similar segmentation accuracies compared to original data, which demonstrated that segmentation models can be developed based on PSF.
|
|
4513.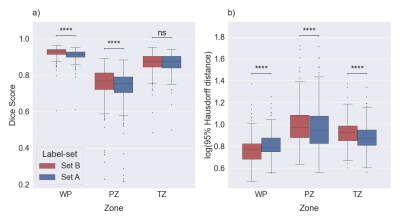 |
83 |
Impact of Label-Set on the Performance of Deep Learning-Based
Segmentation of the Prostate Gland and Zones on T2-Weighted MR
Images
Jakob Meglič1,2,
Mohammed R. S. Sunoqrot1,3,
Tone F. Bathen1,3,
and Mattijs Elschot1,3
1Department of Circulation and Medical Imaging, Norwegian University of Science and Technology (NTNU), Trondheim, Norway, 2Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia, 3Department of Radiology and Nuclear Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway Keywords: Machine Learning/Artificial Intelligence, Prostate Prostate segmentation is an essential step in computer-aided diagnosis systems for prostate cancer. Deep learning (DL)-based methods provide good performance for prostate segmentation, but little is known about the impact of ground truth (manual segmentation) selection. In this work, we investigated these effects and concluded that selecting different segmentation labels for the prostate gland and zones has a measurable impact on the segmentation model performance. |
|
4514.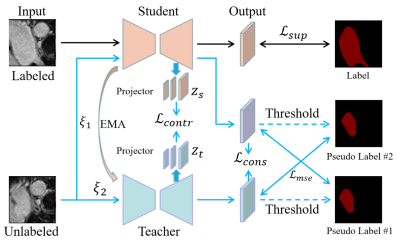 |
84 |
Semi-supervised segmentation for 3D medical image based on
contrast learning
Zhengyong Huang1,2,
Na Zhang1,
Dong Liang1,
Xin Liu1,
Hairong Zheng1,
and Zhanli Hu1
1Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University of Chinese Academy of Sciences, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Data Processing, MRI medical segmentation Semi-supervised segmentation, using large amounts of unlabeled data and small amounts of labeled data, has achieved great success. This paper proposes a semi-supervised segmentation method based on consistent learning and contrast learning. It mainly uses a mean-teacher framework to add consistency losses and contrast losses based on multiscale features to minimize the distance of model responses under different disturbance inputs. In addition, mean square error loss was used to alternately minimize the gap between the teacher and student models. In 3D left atrium data, a Dice coeffivient of 0.8970 was obtained, which was superior to other methods. |
|
4515.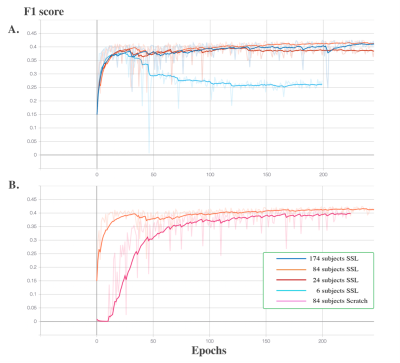 |
85 |
Self-Supervised Learning for Perivascular Spaces segmentation
with enhanced contrast knowledge
Haoyu Lan1,
Arthur W. Toga1,
and Jeiran Choupan1,2
1University of Southern California, Los Angeles, CA, United States, 2NeuroScope Inc., Scarsdale, NY, United States Keywords: Machine Learning/Artificial Intelligence, Segmentation, self-supervised learning Due to the absence of isotropic T2w modality in clinical datasets, it is challenging to enhance the PVS contrast using multiple neuroimage modalities. To overcome this issue, in this work we introduced using self-supervised pre-trained model in the enhanced PVS contrast image space to improve the downstream model segmentation performance when solely using T1w as the training data. The experiment results showed that the proposed method increased segmentation accuracy compared to the model trained from scratch using T1w modality and resulted in faster training and less required training data volume. |
|
4516.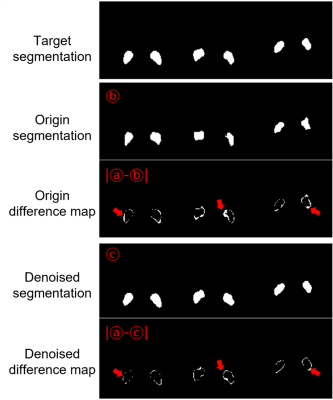 |
86 |
Improved segmentation of neuromelanin region for low SNR short
scan sandwichNM imaging
Joonhyeok Yoon1,
Juhyung Park1,
Yoonho Nam2,
Chul-Ho Sohn3,4,
Junghwa Kang2,
Jonghyo Youn1,
Sooyeon Ji1,
Chungseok Oh1,
and Jongho Lee1
1Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of, 2Department of Biomedical Engineering, Hankuk University of Foreign Studies, Gyeonggi-do, Korea, Republic of, 3Institute of Radiation Medicine, Seoul National University Medical Research Center, Seoul, Korea, Republic of, 4Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of Keywords: Machine Learning/Artificial Intelligence, Parkinson's Disease, Neuromelanin, Denoising Neuromelanin (NM) has been considered an associated biomarker of Parkinson’s disease (PD). Conventional NM visualizeing techniques requires about 5~10min which is sub-optimal for scanning PD patient with movement disorders. Recently, SandwichNM is reduced scan time to 5m 30s, but it is still may not enough. In this research, we approach this issue in the viewpoint of image post processing with denoising techniques to reduce scan time. After 162 NM MRI data acquisition with short scan time, we demonstrated that using denoising technique can improve the distinguishability for NM segmentation. |
|
4517.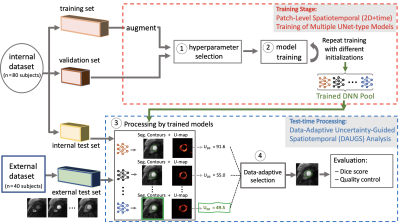 |
87 |
Improved Robustness for Deep Learning-based Segmentation of
Perfusion CMR Using Data Adaptive Uncertainty-guided
Spatiotemporal Analysis
Dilek Mirgun Yalcinkaya1,2,
Khalid Youssef3,
Bobak Heydari4,
Subha Raman3,5,
Rohan Dharmakumar3,5,
and Behzad Sharif1,3,5
1Laboratory for Translational Imaging of Microcirculation, Indiana University (IU) School of Medicine, Indianapolis, IN, United States, 2Elmore Family School of Electrical and Computer Engineering, Purdue University, West Lafayette, IN, United States, 3Krannert Cardiovascular Research Center, IU School of Medicine/IU Health Cardiovascular Institute, Indianapolis, IN, United States, 4Stephenson Cardiac Imaging Centre, Department of Cardiac Sciences, University of Calgary, Calgary, AB, Canada, 5Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States Keywords: Machine Learning/Artificial Intelligence, Segmentation We proposed and validated Data Adaptive Uncertainty-Guided Spatiotemporal (DAUGS) analysis that leverages the data-driven uncertainty map of the segmentation contours among a pool of trained deep neural networks (DNNs) and automatically selects the segmentation result with the highest level of certainty. Our results suggest that proposed DAUGS and standard DNN-based analysis demonstrated on-par performance on the internal test set which is from the same institution as training set and acquired with FLASH sequence. In contrast, DAUGS analysis considerably outperformed DNN-based analysis on the external test set which was acquired with a bSSFP pulse sequence at a different institution, demonstrating the improved robustness of the proposed method despite limited training data. |
|
4518.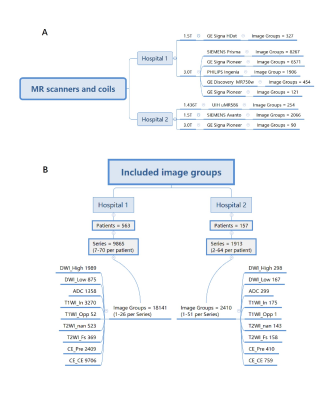 |
88 |
AI algorithms for classification of the mpMRI image sequences
and segmentation of the prostate gland: an external validation
study
Kexin Wang1
1Capital Medical University, Beijing, China Keywords: Prostate, Machine Learning/Artificial Intelligence In this study we evaluate the generalization of the AI algorithms for the classification of the mpMRI image sequences and the segmentation of the prostate gland with multicenter external dataset. A total of 719 patients who underwent multiparametric MRI (mpMRI) of the prostate were collected retrospectively from two hospitals. AI algorithms were tested for classification of the image type and segmentation of the prostate gland. The AI models demonstrated good performance in the external validation in the task of image classification and prostate gland segmentation. |
|
4519.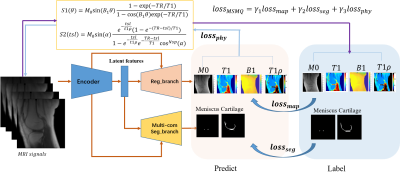 |
89 |
Reduced input combination study for the Simultaneous
Multi-Tissue Segmentation and Multi-Parameter Quantification
Network (MSMQ-Net) of Knee
Xing Lu1,
Yajun Ma1,
Jiyo Athertya1,
Chun-Nan Hsu2,
Eric Y Chang1,3,
Amilcare Gentili1,3,
Christine Chung1,
and Jiang Du1
1Department of Radiology, University of California, San Diego, San Diego, CA, United States, 2Department of Neurosciences, University of California, San Diego, San Diego, CA, United States, 3Radiology Service, Veterans Affairs San Diego Healthcare System, San Diego, CA, United States Keywords: Osteoarthritis, Quantitative Imaging To accelerate ultrashort echo time (UTE) based multi-parameter quantitative MRI (qMRI), we performed comparison studies on the input MRI image numbers and the effects on the prediction quality of the MSMQ-Net. The MSMQ-net was modified and trained accordingly with different combinations of inputs. Both image similarity and regional analysis were evaluated. The results demonstrate that 90% accuracy can be achieved for both UTE-T1 and UTE-T1rho mapping when the scan time is reduced by 75%. |
|
4520.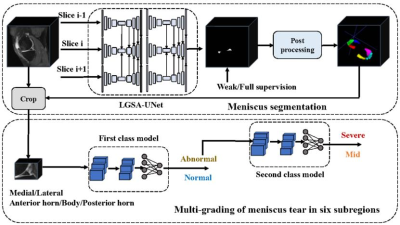 |
90 |
Full and weak supervision networks for meniscus segmentation and
multi classification based on MRI: data from the Osteoarthritis
Initiative
Kexin Jiang1,
Yuhan Xie2,
Zhiyong Zhang2,
Jiaping Hu1,
Shaolong Chen2,
Zhongping Zhang3,
Changzhen Qiu2,
and Xiaodong Zhang1
1Department of Medical Imaging, The Third Affiliated Hospital of Southern Medical University, GuangZhou, China, 2Electronics and Communication Engineering, Sun Yat-sen University, GuangZhou, China, 3Philips Healthcare, GuangZhou, China Keywords: Joints, Joints, meniscus Quantitative MRI of meniscus morphology (such as MOAKS system) have shown clinical relevance in the diagnosis of osteoarthritis. However, it requires a large workload, and often lead to deviation due to reader’s subjectivity. Therefore, based on automated segmentation of six horns of meniscus using fully and weakly supervised networks, we established two-layer cascaded classification models that can detect the meniscal lesions and further classify them into three types, and finally achieved excellent performance. This can improve the efficiency and accuracy of using quantitative MRI to study KOA in the future. |
|
4521.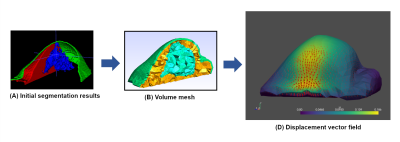 |
91 |
Finite element simulation of breast hyperelasticity to develop a
non-rigid registration model by deep learning
Fares OUADAHI1,
Aurélie PETER1,
Anaïs BERNARD1,
and Julien ROUYER1
1Research and Innovation Department, Olea Medical, La Ciotat, France Keywords: Breast, Data Processing, Registration, Motion correction This work is part of an ongoing study to correct accidental breast motion during dynamic contrast-enhanced imaging. Here, we explored a finite element method to generate “moving” images based on a “fixed” one. The proposed methodology can help in the constitution of realistic dataset with large variety of motion to support the development of a dedicated non-rigid registration model by deep learning. |
|
4522.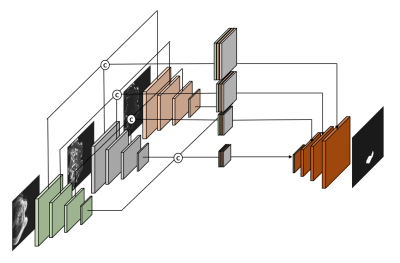 |
92 |
Ensemble Multi-Path U-net Segmentation Algorithm for Breast
Lesion based on Multi-Modality Image
Hang Yu1,
Zichuan Xie2,
Lizhi Xie3,
Zhiheng Liu1,
Lina Zhang4,
Siyao Du4,
Xiangjie Yin1,
Chenyang Li1,
Wenhong Jiang4,
Yuru Guo1,
and Zhongqi Kang4
1School of Aerospace Science and Technology, Xidian University, Xi'an, China, 2Guangzhou institute of technology, Xidian University, Guangzhou, China, 3GE Healthcare, Beijing, China, 4Department of Radiology, The First Hospital of China Medical University, Shenyang, China Keywords: Breast, Machine Learning/Artificial Intelligence, Deep Learning The multimodal MRI data is often ultilized for breast cancer analysis, and by now still difficult and inefficient to explored by segmentation algorithms. In this paper, we propose a MP-Unet based on U-net convolutional neural network, which can effectively ensemble multiple inputs of modal data and obtain accuracy segmentation results at the end. In MP-Unet, we reused some good quality modal data for training. The multiple MP-Unet models are further integrated based on Bagging algorithm to improve the segmentation accuracy of lesions. Experiments suggest that our proposed method has a huge performance improvement. |
|
4523. |
93 |
Volumetric vocal tract segmentation using a deep transfer
learning 3D U-Net model
Subin Erattakulangara1,
Sarah Gerard1,
Karthika Kelat1,
Katie Burnham2,
Rachel Balbi2,
David Meyer2,
and Sajan Goud Lingala1,3
1Roy J Carver Department of Biomedical Engineering, University of Iowa, iowa city, IA, United States, 2Janette Ogg Voice Research Center, Shenandoah University, Winchester, IA, United States, 3Department of Radiology, University of Iowa, iowa city, IA, United States Keywords: Machine Learning/Artificial Intelligence, Head & Neck/ENT The investigation into the 3D airway area is the prerequisite step for quantitatively studying the anatomical structures and function of the upper airway. Segmentation of upper airway can be considered as one of the stepping stones for this investigation. In this work, we propose a transfer learning-based 3D U-Net model with a ResNet encoder for vocal tract segmentation with small datasets training. We demonstrate its utility on sustained volumetric vocal tract MR scans from the recently released French speaker database. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.