Digital Poster
ML/AI for Data Synthesis, Generative Models & Quantitative MRI II
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
5179.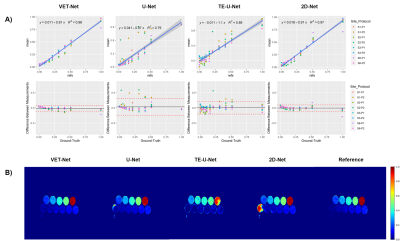 |
81 |
Reproducible DL-based approach for liver PDFF quantification
Juan Pablo Meneses1,2,3,
Cristobal Arrieta1,2,
Pablo Irarrazaval1,3,4,
Cristian Tejos1,3,
Marcelo Andía1,2,5,
Carlos Sing Long1,4,6,
and Sergio Uribe1,2,5
1Biomedical Imaging Center, Pontificia Universidad Católica de Chile, Santiago, Chile, 2i-Health Millennium Institute for Intelligent Healthcare Engineering, Santiago, Chile, 3Department of Electrical Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 4Institute for Biological and Medical Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 5Radiology Department, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile, 6Institute for Mathematical & Computational Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile Keywords: Machine Learning/Artificial Intelligence, Quantitative Imaging, Convolutional Neural Network Liver PDFF is a biomarker correlated with hepatic pathologies. Recently, several Deep Learning (DL) methods have been proposed to accelerate the necessary post-processing to estimate PDFF. However, none of these techniques had been assessed in terms of bias and precision, as suggested by the ISMRM quantitative MR study group. We propose a two-stages framework denoted Variable Echo Times neural Network (VET-Net), which considers multi-echo MR images and their echo times to estimate PDFF. VET-Net showed a bias of -1.35% when tested over a multi-site phantom dataset, and a within-standard deviation of 0.81% over liver MR images with different TEs. |
|
5180.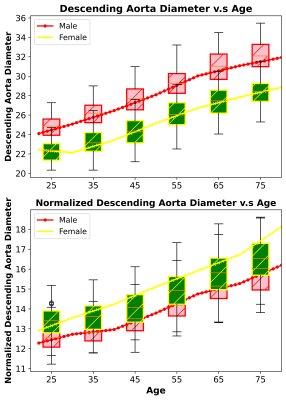 |
82 |
Automatic Aorta Quantification and Aneurysm Detection using 3D
Deep Learning on Large-scale Non-Cardiac-Gated MRI
Thanh-Duc Nguyen1,
Saurabh Garg1,
Nasrin Akbari1,
Saqib Basar1,
Sean London2,
Yosef Chodakiewitz2,
Rajpaul Attariwala1,2,
and Sam Hashemi1,2
1Voxelwise Imaging Technology Inc., Vancouver, BC, Canada, 2Prenuvo, Vancouver, BC, Canada Keywords: Machine Learning/Artificial Intelligence, Cardiovascular, Aorta quantification We present a validated AI technique for quantification of whole aorta diameter from non-cardiac-gated MRI. The mean absolute error between AI-predicted and radiologist-reported diameter is less than 0.27 and 0.249 cm, while Pearson correlation is 0.72 for the ascending (p-value=0.0002) and 0.74 for the descending aorta (p-values=0.0012). SVM classifiers indicated that a 3.15 cm (AUC=0.93) threshold is used to detect a descending aortic aneurysm while a 4.3 cm (AUC=0.92) threshold is used for ascending aortic aneurysm. Large-scale analysis on 3485 individuals showed aortic diameter to increase with age in male and female populations. |
|
5181.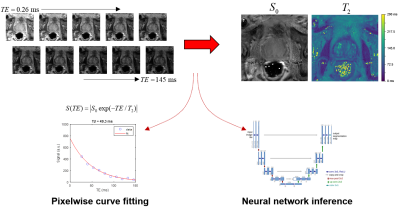 |
83 |
Training Strategies for Convolutional Neural Networks in
Prostate T2 Relaxometry
Patrick Bolan1,
Sara Saunders1,
Mitchell Gross1,
Kendrick Kay1,
Mehmet Akcakaya2,
and Gregory Metzger1
1Center for MR Research / Radiology, University of Minnesota, Minneapolis, MN, United States, 2Electrical and Computer Engineering, University of Minnesota, Minneapolis, MN, United States Keywords: Machine Learning/Artificial Intelligence, Prostate, Relaxometry This work uses convolutional neural networks (CNNS) with two training strategies for estimating quantitative T2 values from prostate relaxometry measurements, and compares the results to conventional non-linear least squares fitting. The CNN trained with synthetic data in a supervised manner gave lower median errors and better noise robustness than either NLLS fitting or a CNN trained on in vivo data with a self-supervised loss. |
|
5182.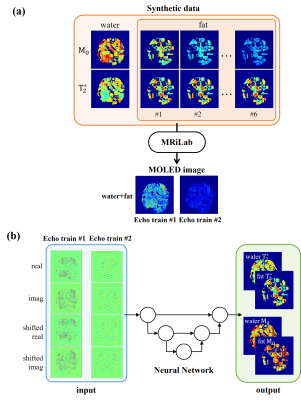 |
84 |
Single-shot water/fat separation T2* mapping with multiple
overlapping-echo detachment imaging
Qing Lin1,
Weikun Chen1,
Jian Wu1,
Taishan Kang2,
Xinran Chen1,
Zhigang Wu3,
Shuhui Cai1,
and Congbo Cai1
1Department of Electronic Science, Xiamen University, Xiamen, China, 2Magnetic Resonance Center, Zhongshan Hospital Afflicated to Xiamen University, Xiamen, China, 3MSC Clinical & Technical Solutions, Philips Healthcare, Shenzhen, China Keywords: Machine Learning/Artificial Intelligence, Quantitative Imaging, water/fat separation Most of the water/fat separation techniques need to acquire multiple images with different echo time, which usually take long acquisition time. Multiple overlapping-echo detachment (MOLED) imaging can shorten acquisition time by acquiring multiple MR echo signals in the same k-space. Here, a new method for fast water/fat separation T2* mapping with MOLED technology was proposed, which can obtain quantitative T2* maps and M0 maps of water and fat in a single shot. In vivo experiment demonstrates that the proposed method can obtain accurate quantitative parameter values and accurate separation of water and fat images. |
|
5183.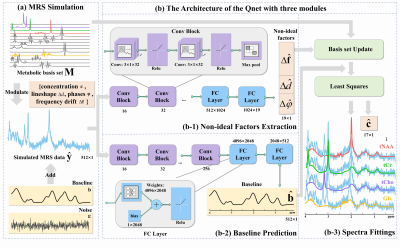 |
85 |
Deep Learning Quantification of Magnetic Resonance Spectroscopy
Based on Basis set and Exponential Priors
Dicheng Chen1,
Huiting Liu1,
Yirong Zhou1,
Xi Chen2,
Zhangren Tu1,
Liangjie Lin3,
Zhigang Wu3,
Jiazheng Wang3,
Di Guo4,
Jianzhong Lin5,
and Xiaobo Qu1
1Department of Electronic Science, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2McLean Hospital, Harvard Medical School, Belmont, MA, United States, 3Philips Healthcare, Beijing, China, 4School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, 5Department of Radiology, The Zhongshan Hospital affiliated to Xiamen University, Xiamen, China Keywords: Machine Learning/Artificial Intelligence, Brain, Magnetic Resonance Spectroscopy, Quantification Quantification of 1H-MRS is difficult because of the overlapping of individual metabolite signals, non-ideal acquisition conditions, and strong background signal interference. We introduced Deep Learning (DL) method to learn these effects to improve the accuracy of the quantification. Results indicate that, compared with the conventional method LCModel, the proposed Qnet (Quantification deep learning network) shows better quantification for both simulated and in vivo acquired MRS data with lower fitting errors and enhanced stability. |
|
5184.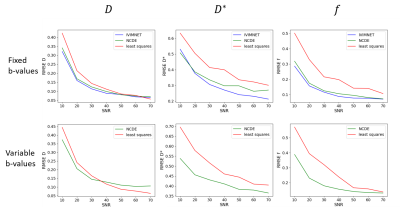 |
86 |
Quantitative MRI Parameter Estimation using Neural Controlled
Differential Equations: Proof-of-Concept in Intra-voxel
Incoherent Motion
Daan Kuppens1,
Daisy van den Berg1,
Sebastiano Barbieri2,
Aart J. Nederveen1,
and Oliver J. Gurney-Champion1
1Radiology & Nuclear Medicine, Amsterdam UMC, Amsterdam, Netherlands, 2Centre for Big Data Research in Health, University of New South Wales Sydney, Sydney, Australia Keywords: Machine Learning/Artificial Intelligence, Quantitative Imaging In quantitative MRI, tissue properties are estimated from MRI data using bio-physical models that relate the MRI signal to the underlying tissue properties via model parameters. Deep learning can improve parameter estimation, but is conventionally dependent on the input being either a fixed set of input signals or a series of regularly sampled signals. Neural controlled differential equations (NCDEs) are models that are independent of the configuration of input data. NCDEs have similar performance to state-of-the-art acquisition-specific deep learning methods in estimating intra-voxel incoherent motion parameters. Therefore, NCDEs are a generic purpose tool for parameter estimation in quantitative MRI. |
|
5185.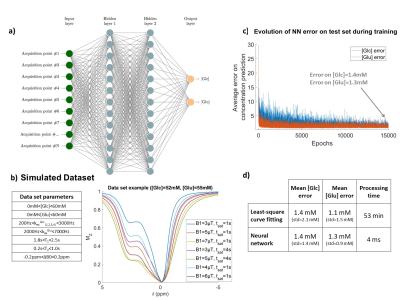 |
87 |
Deep-learning and feature selection for fast, quantitative and
specific CEST imaging
Cecile Maguin1 and
Julien Flament1
1Université Paris-Saclay, CEA, CNRS, MIRCen, Laboratoire des Maladies Neurodégénératives, Fontenay-aux-roses, France Keywords: Machine Learning/Artificial Intelligence, CEST & MT, Feature selection We propose an artificial neural network combined with a feature selection scheme for fast, quantitative CEST imaging, designed for specificity. Our NN was evaluated on glucose phantoms and glutamate/glucose mixed phantoms and goes beyond performances of classical fittings approaches. |
|
5186.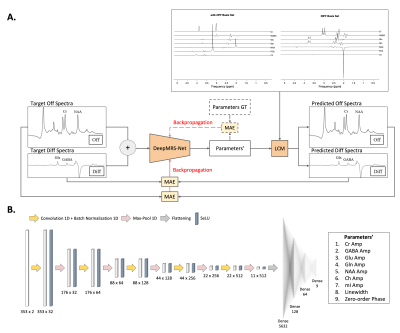 |
88 |
DeepMRS-Net: QUANTIFICATION OF MAGNETIC RESONANCE SPECTROSCOPY
MEGA-PRESS DATA USING DEEP LEARNING
Christopher Jiaming Wu1 and
Jia Guo2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia University, New York, NY, United States Keywords: Machine Learning/Artificial Intelligence, Brain, Convolutional Neural Networks, Multi-class Regression, Unsupervised Learning Quantification of metabolites in the human brain in vivo from magnetic resonance spectra (MRS) has many applications in medicine and psychology, but it remains a challenging task despite considerable research efforts. In this paper, we propose quantification of metabolites from MEGA-PRESS data using deep learning through an unsupervised learning approach. A regression framework based on the Convolutional Neural Networks (CNN) is introduced for estimation of spectral parameters including the relative concentrations of metabolites, line-broadening, and zero-order phase. The results show that the model is capable of reliably fitting in vivo data. |
|
5187.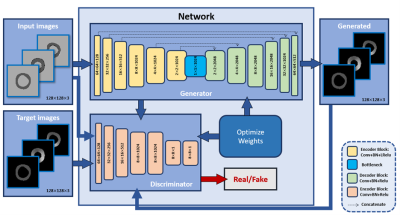 |
89 |
Myocardial strain generation from cine MR images using an
automated deep learning network
Dayeong An1 and
El-Sayed Ibrahim1
1Biomedical Engineering, Medical College of Wisconsin, Milwaukee, WI, United States Keywords: Machine Learning/Artificial Intelligence, Heart Current gold-standard method for obtaining myocardial strain is based on MRI tagged images, although this increases the MRI exam time and requires special analysis software. We propose to use cine MR images to train a deep neural-network to generate myocardial strain based on target strain maps generated from tagged images acquired at the same locations and timepoints as the cine images. The results showed high agreement between the output and target strain maps. Our method not only saves MRI scan time by acquiring only cine images but also pre and post image processing time by quantifying myocardial strains automatically. |
|
5188.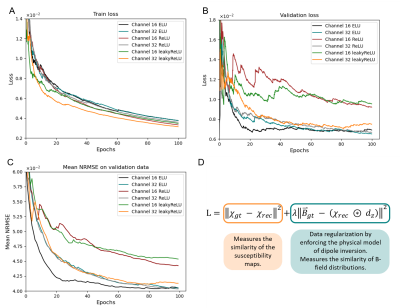 |
90 |
Comparison of activation functions for optimizing deep learning
models solving QSM-based dipole inversion
Simon Graf1,
Nora Küchler1,
Walter Wohlgemuth1,
and Andreas Deistung1
1University Hospital Halle (Saale), Halle (Saale), Germany Keywords: Machine Learning/Artificial Intelligence, Quantitative Susceptibility mapping Deploying deep learning models for quantitative susceptibility mapping is driven by optimizing hyper-parameters and using suitable architectures. We investigated the impact of activation functions on network model training. ELU-, leaky ReLU- and ReLU-models with 16 and 32 initial channels were tested for solving dipole inversion on synthetic susceptibility data. All models showed convergence after completing 100 training epochs. However, the 16-channel-ELU-model achieved low losses after only 20 training epochs and showed similar reconstruction performance to the 32-channel-ELU-model. Using the ELU activation allows the use of smaller network models resulting in fewer memory requirements and less training time. |
|
5189.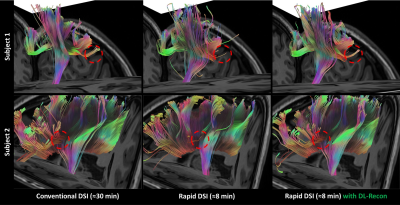 |
91 |
8-minute Rapid Whole-brain Diffusion Spectral Imaging with Deep
Learning-based Reconstruction: A Feasibility Study
Yuhui Xiong1,
Xiaocheng Wei1,
Jiankun Dai1,
Yang Fan1,
Jie Lu2,
and Bing Wu1
1GE Healthcare MR Research, Beijing, China, 2Department of Radiology, Xuanwu Hospital, Capital Medical University, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence This study aims to shorten the diffusion spectral imaging (DSI) scan time to a clinically acceptable level while providing satisfactory complex white matter fiber structure description as well as accurate diffusion metric quantification by applying deep learning-based reconstruction. Images were acquired using conventional (≈30 min) and rapid (≈8 min) DSI sequences, and reconstructed using conventional and DL-based methods, respectively. Atlas-based fiber-tracking and diffusion metrics quantification from various advanced models were conducted. The results demonstrated that the 8-minute rapid DSI sequence combined with DL-recon can provide complex fiber structure tractography and advanced diffusion metric quantification of satisfactory quality. |
|
5190.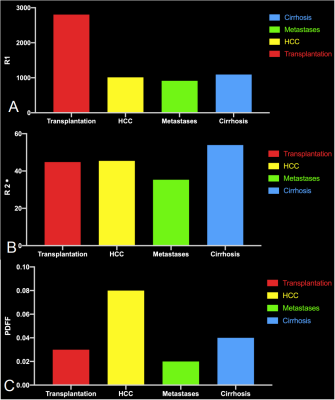 |
92 |
Comparison of R1, R2* and PDFF mapping by simultaneous
multi-relaxation-time Imaging (TXI) method in hepatic disease
Yishuang Wang1,
Tianxiang Huang1,
Meining Chen2,
XU YAN2,
and Longlin Yin1
1Radiology, Sichuan Provincial People's Hospital, Chengdu, China, 2MR Scientific Marketing, Siemens Healthcare, Shanghai, China Keywords: Machine Learning/Artificial Intelligence, Liver MR multiparametric quantitative techniques have an important role in liver-related diseases.We achieved simultaneous proton density fat fraction(PDFF), R2*and R1 quantification of different liver diseases using a simultaneous multi-relaxation-time Imaging(TXI) technique.We observed differences of PDFF,R2* and R1 mapping between healthy volunteers and patients with cirrhosis,HCC,metastatic carcinoma of liver,and liver transplantation.Rapid scanning increases usage of this method in the clinic.These quantitative values can be biomarkers for assessing liver function and liver tissue characterization. |
|
5191.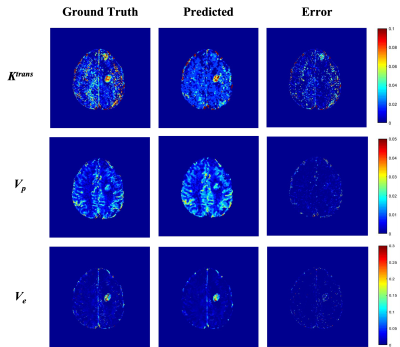 |
93 |
Quantitative DCE-MRI parameter estimation using Deep Learning
Framework in the Brain Tumor Patients
Piyush Kumar Prajapati1,
Rakesh Kumar Gupta2,
and Anup Singh1,3,4
1Centre for Biomedical Engineering, Indian Institute of Technology Delhi, New Delhi, India, 2Department of Radiology, Fortis Memorial Research Institute, Gurugram, India, 3Yardi School of Artificial Intelligence, Indian Institute of Technology Delhi, New Delhi, India, 4Department of Biomedical Engineering, All India Institute of Medical Sciences, New Delhi, India Keywords: Machine Learning/Artificial Intelligence, Brain Tracer Kinetic (TK) parametric maps are obtained from Dynamic Contrast Enhanced (DCE) - MRI which aid in the detection and grading of brain tumors. Conventionally, TK maps are obtained using Non-Linear-Least-Square (NLLS) fitting approach, which is time consuming and data noise. In the current study, we implemented a deep learning framework whose backbone is attention networks to estimate TK parametric maps. Transfer learning was performed to extend work from synthetic data to high-grade glioma (HGG) patients’ DCE-MRI data to obtain better quality TK maps in lesser time. |
|
5192.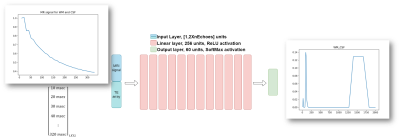 |
94 |
Recovery of T2 distribution from quantitative T2-weighted MRI
with physically-driven deep learning
Hadas Ben-Atya1 and
Moti Freiman1
1Faculty of Biomedical Engineering, Technion, Haifa, Israel Keywords: Machine Learning/Artificial Intelligence, Relaxometry Recovery of T2 distribution of tissue from MRI data acquired at multiple echo times has the potential to be used as a biomarker for the assessment of various pathologies, including stroke and epilepsy, investigation of neurodegenerative diseases, and tumor characterization. Current deep neural networks (DNN) for T2 distribution recovery are highly sensitive to variations in the acquisition parameters such as different echo times. We present a new physically-driven DNN model that encodes the TE acquisition parameters as part of its architecture. Our model accurately recovers the T2 distribution, regardless of variations in SNR and in the acquisition parameters. |
|
5193.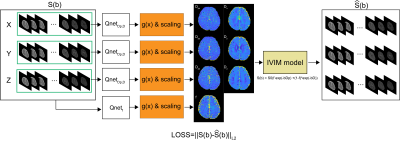 |
95 |
Constraint function for IVIM quantification using unsupervised
learning
Wonil Lee1,
Beomgu Kang1,
Jongyeon Lee1,
Georges El Fakhri2,
Chao Ma2,
Yeji Han3,
Jun-Young Chung4,
Young Noh5,
and HyunWook Park1
1The School of Electrical Engineering, Korea Advanced Institute of Science and Technology (KAIST), Yuseong-gu, Korea, Republic of, 2Massachusetts General Hospital, Boston, MA, United States, 3Department of Biomedical Engineering, Gachon University, Incheon, Korea, Republic of, 4Department of Neuroscience, College of Medicine, Gachon University, Incheon, Korea, Republic of, 5Department of Neurology, Gil Medical Center, Gachon University College of Medicine, Incheon, Korea, Republic of Keywords: Machine Learning/Artificial Intelligence, Quantitative Imaging, Intravoxel incoherent motion Recently, various methods have been proposed to quantify intravoxel incoherent motion parameters. Many studies have shown that quantification methods using deep learning can accurately estimate IVIM parameters. Unsupervised learning is useful when quantifying IVIM parameters for in-vivo data because it does not require label data. However, in some cases, loss function does not converge as iteration increases. Constraint functions can be used to solve these problems by limiting the range of estimated outputs. In this study, we investigated the effects of constraint function to limit the range of estimated output. |
|
5194.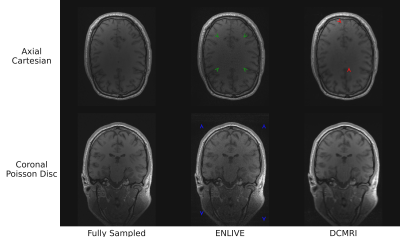 |
96 |
Evaluating different k-space undersampling schemes with
iterative and deep learning image reconstruction for fast
multi-parameter mapping
Kornelius Podranski1,
Kerrin J. Pine1,
Timoteo Colnaghi2,
Andreas Marek2,
Patrick Scheibe1,
Nico Scherf1,3,
and Nikolaus Weiskopf1,4
1Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 2Max Planck Computing and Data Facility, Garching (Munich), Germany, 3Neural Data Science and Statistical Computing, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 4Felix Bloch Institute for Solid State Physics, Faculty of Physics and Earth Sciences, Leipzig University, Leipzig, Germany Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, Brain Approaches for accelerating multi-echo gradient echo (ME-GRE) acquisitions as a basis for multi-parameter mapping (MPM) were explored. Fully sampled ME-GRE data were retrospectively undersampled to equispaced Cartesian, CAIPIRINHA and Poisson disc patterns. Echoes were jointly reconstructed with the iterative ENLIVE algorithm and the machine learning/artificial intelligence adapted DeepcomplexMRI (DCMRI) approach. The approaches result in comparable peak signal-to-noise ratio (PSNR) and structural similarity index measure (SSIM), but show different types and different levels of artifacts. The DCMRI approach promises fast reconstruction and flexibility in the choice of undersampling patterns for ME-GRE imaging in the future. |
|
5195.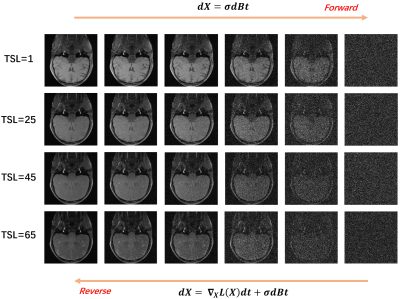 |
97 |
Joint Distribution Modeling for Accelerated T1rho Reconstruction
Congcong Liu1,
Zhuo-Xu Cui1,
Yuanyuan Liu1,
Chentao Cao1,
Jing Cheng1,
Yanjie Zhu1,
Haifeng Wang1,
and Dong Liang1,2
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Pazhou Lab, Guangzhou, China Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction Traditional hand-craft designed methods to accelerated T1rho mapping have limited characterisation capabilities, while deep learning methods lack the interpretability. On the other hand, the joint distribution is the most direct and accurate way to characterize the correlation between different images. Therefore, we attempt to propose a joint distribution estimation method and use it to construct a T1$$$\rho$$$ reconstruction model. In particular, we use a score-based diffusion model to model the joint distribution of acquired T1rho-weighted images. Moreover, the corresponding reconstruction model is solved using the Langevin gradient descent method. Finally, numerical experiments validate the effectiveness of the proposed method. |
|
5196.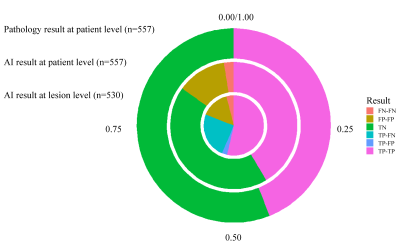 |
98 |
A multicenter external validation study of the role of AI
algorithm in detecting and localizing clinically significant
prostate cancer on mpMRI
Zhaonan Sun1,
Xiaoying Wang2,
and Kexin Wang3
1Department of Radiology, Peking University First Hospital, Beijing, China, 2Department of Radiology, Peking University First Hospital, Beijing, China, 3School of Basic Medical Sciences, Capital Medical University, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Prostate A total of 557 mpMRI data were retrospectively collected from three hospitals to build an external validation dataset, with 245 csPCa cases and 312 non-csPCa cases. The csPCa lesions were annotated based on pathology records by two experienced radiologists. AI algorithms were used to automatically detect and localize the suspicious csPCa areas on the T2WI and ADC maps. The metrics of sensitivity, specificity, and accuracy were used to evaluate the diagnostic efficacy of the AI algorithms at the lesion level, the sextant level, and the patient level. |
|
5197.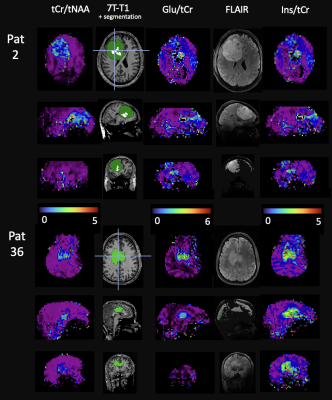 |
99 |
Glioma Classifications with 7T MR Spectroscopic Imaging
Sukrit Sharma1,
Cornelius Cadrien2,
Philipp Lazen1,
Hangel Gilbert1,
Roxane Licandro3,
Wolfgang Bogner1,
and Georg Widhalm2
1High-field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, Medical University of Vienna, Vienna, Austria, 2Medical University of Vienna, Vienna, Austria, 3Department of Biomedical Imaging and Image-guided Therapy, Computational Imaging Research Lab (CIR) and Laboratory for Computational Neuroimaging, A.A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital / Harvard Medical School, Charlestown, MA, US., Medical University of Vienna, Vienna, Austria Keywords: Machine Learning/Artificial Intelligence, Brain, Glioma To contribute to better tumour classification and thus enhancing patient outcomes, we statistically analysed metabolic maps of 37 glioma patients obtained using high resolution 7T MRSI. We tested and optimised different semi-supervised learning based classification approaches. Random forest classification of IDH mutation status and tumour grade in clinical imaging based segmented tumour regions yielded high diagnostic accuracy with AUC of 86% and 99% respectively. We found Glu, Gln, GSH, tCho, Ins, Gly and tCr as important determining features. These are similar to comparable SVS studies while providing the advantage of whole-brain coverage. |
|
5198.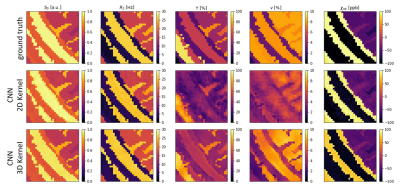 |
100 |
3D CNN for Oxygen Extraction Fraction Mapping with combined QSM
and qBOLD
Patrick Kinz1 and
Lothar R Schad1
1Computer Assisted Clinical Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany Keywords: Machine Learning/Artificial Intelligence, Oxygenation We developed a CNN for OEF mapping from QSM+qBOLD data, which utilizes utilizes 3D convolutional layers. Two dimensions for the spatial components of an image and one dimension for the temporal component in the qBOLD data. The results are an improvement over our previous 2D CNN, but even the more advanced network architecture struggles with voxels, that have a very low deoxyhemoglobin content. In this abstract we also study with simulated data when the CNN produces reliable results and when it predicts default values for the reconstructed parameters instead. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.