Digital Poster
ML/AI for General Image Analysis & Post-Processing II
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
4907.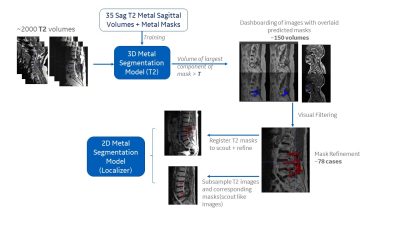 |
141 |
Automatic localization of metal artifacts regions on MRI scout
images
Deepa Anand1,
Dattesh Shanbhag1,
Chitresh Bhushan2,
Kavitha Manickam2,
and RAdhika Madhavan2
1GE Healthcare, Bangalore, India, 2GE Healthcare, Niskayuna, NY, United States Keywords: Machine Learning/Artificial Intelligence, Artifacts, Metal implants In MRI, presence of metal can cause blooming artifacts on MRI images. Automatically detecting the presence of metal and localizing it on the scout images itself can help streamline the MRI imaging workflow decisions downstream. In this work, we demonstrate a DL framework for hot spotting of metal/metal affected regions based on 2D three-plane MRI scout images of spine. The results indicate that the solution not only detects metal regions well on spine images (on which it was trained), but also is generalizable enough to work on other anatomies such as knee which was not part of the training data. |
|
4908.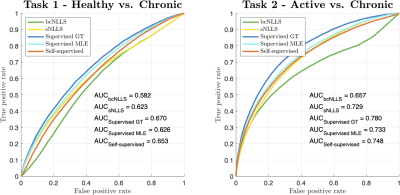 |
142 |
Do deep learning-based qMRI parameter estimators improve
clinical task performance?
Sean C. Epstein1,2,
Timothy J. P. Bray3,4,
Margaret Hall-Craggs3,4,
and Hui Zhang1,2
1Computer Science, UCL, London, United Kingdom, 2Centre for Medical Image Computing, UCL, London, United Kingdom, 3Centre for Medical Imaging, UCL, London, United Kingdom, 4Imaging, UCLH, London, United Kingdom Keywords: Machine Learning/Artificial Intelligence, Data Analysis We compare modern deep learning (DL)-based parameter-estimation methods to their traditional maximum-likelihood estimation (MLE) counterparts by evaluating each approach’s performance in two clinical classification tasks. This is motivated by recent work demonstrating the inherent bias-variance trade-off that differentiates different DL-based approaches. Results show how these trade-offs manifest in the ‘real world’ of tissue classification, and how they compare to the performance achievable with conventional iterative MLE. |
|
4909.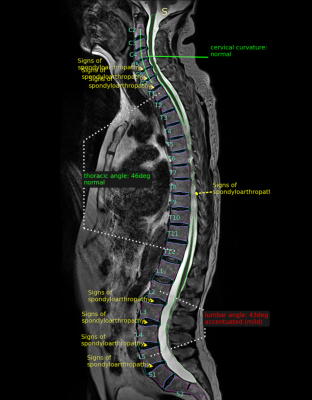 |
143 |
Quantitative Assessment of the Whole Spine in T2 MRI Using Deep
Learning
Siavash Khallaghi1,
Lucas Porto1,
Sean London2,
Yosef Chodakiewitz2,
Rajpaul Attariwal1,
and Sam Hashemi1
1Voxelwise Imaging Technology Inc., Vancouver, BC, Canada, 2Prenuvo, Vancouver, BC, Canada Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Spine, spondyloarthropathy, spondylosis We present a fully automatic system for the quantitative assessment of discs and vertebrae using convolutional neural networks. The proposed algorithm works in three stages: 1) segmentation/identification of spinal anatomy; 2) curvature analysis; and 3) detection of pathological conditions of intervertebral discs. We validate the proposed approach on a large dataset of 1,500 subjects with sagittal T2-weighted whole spine MRI, obtained as part of a whole body MRI protocol in a preventative health screening program. |
|
4910.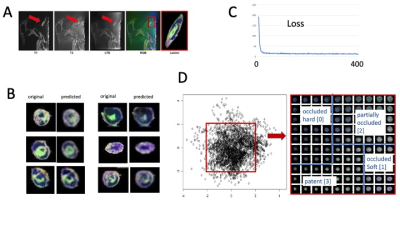 |
144 |
Classification of Plaque Composition in Peripheral Arterial
Disease with Multi-Contrast Histology using 7 Tesla and a
Variational AutoEncoder
Christof Karmonik1,
Lily Buckner2,
Kayla Wilhoit1,
and Trisha Roy3
1Houston Methodist Research Institute, Houston, TX, United States, 2Houston Methodst Research Institute, Houston, TX, United States, 3Houston Methodist Hospital, Houston, TX, United States Keywords: Machine Learning/Artificial Intelligence, High-Field MRI, ultra-short TE, 7 Tesla To differentiate soft versus hard peripheral arterial disease (PAD) plaques, a dedicated ultra-high field magnetic resonance imaging (MRI) histology protocol using three different image contrast (ultra-short TE, T1-weighted and T2-weighted) was used to scan on a clinical 7 Tesla human MRI scanner six amputated legs immediately after surgery that were harboring PAD lesions. A 2D convolutional network (CNN) variational autoencoder (VAE) was created. For each lesion, average classification values along the long axis of the artery were determined from latent space and ranged from partially occluded to patent (three lesions), soft-plaque occluded (two) and hard-plaque occluded (one). |
|
4911.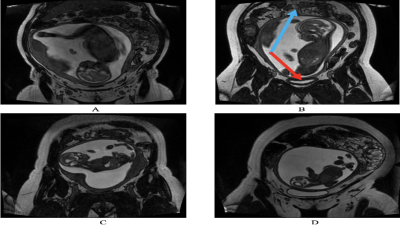 |
145 |
Automatic Fetal Orientation Detection Algorithm in Fetal MRI
Joshua Eisenstat1,
Matthias Wagner2,
Logi Vidarsson3,
Birgit Ertl-Wagner2,4,
and Dafna Sussman1,5,6
1Department of Electrical, Computer and Biomedical Engineering, Faculty of Engineering and Architectural Sciences, Toronto Metropolitan University, Toronto, ON, Canada, 2Division of Neuroradiology, The Hospital for Sick Children, Toronto, ON, Canada, 3Department of Diagnostic Imaging, The Hospital for Sick Children, Toronto, ON, Canada, 4Department of Medical Imaging, Faculty of Medicine, University of Toronto, Toronto, ON, Canada, 5Institute for Biomedical Engineering, Science and Technology (iBEST), Toronto Metropolitan University and St. Michael’s Hospital, Toronto, ON, Canada, 6Department of Obstetrics and Gynecology, Faculty of Medicine, University of Toronto, Toronto, ON, Canada Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Convolutional Neural Networks, Fet-Net Fetal orientation determines the mode of delivery. It is also important for sequence planning in fetal MRI. This abstract proposes Fet-Net, a deep-learning algorithm, which uses a novel convolutional neural network (CNN) architecture, to automatically detect fetal orientation from a 2-dimensional (2D) magnetic resonance imaging (MRI) slice. 6,120 2D MRI slices displaying vertex, breech, oblique and transverse fetal orientations were used for training, validation and testing. Fet-Net achieved an average accuracy and F1 score of 97.68%, and a loss of 0.06828. Fet-Net was able to detect and classify fetal orientation, which may serve to accelerate fetal MRI acquisition. |
|
4912.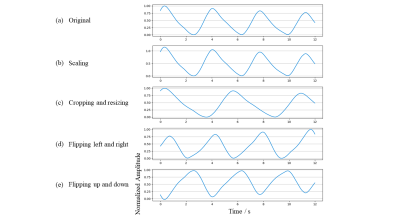 |
146 |
Deep Learning Based Real-time Quality Assessment of Pilot Tone
Respiratory Signals
Huixin Tan1 and
Yantu Huang1
1Siemens Shenzhen Magnetic Resonance Ltd., China, Shenzhen, China Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence Pilot Tone (PT) respiratory signals are susceptible to interference like patient bulk motion or radio frequency interference. The signal curve is prone to bad when remains strong interference after suppression processing in the learning phase, resulting in low triggering accuracy and efficiency. With real-time quality assessment in the learning phase, a good-quality signal can be selected to learn better processing parameters. To ensure robustness and inference time, a tiny CNN is utilized to classify the signal into good and bad quality. Experimental results demonstrate that the proposed method has a strong ability to assess the quality of PT respiratory signals. |
|
4913.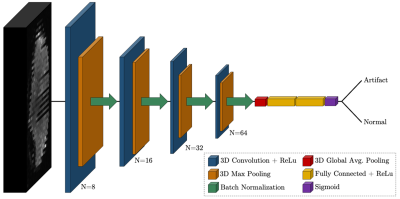 |
147 |
Automated Motion Artifact Detection in Early Pediatric Diffusion
MRI Using a Convolutional Neural Network
Jayse Merle Weaver1,2,
Marissa DiPiero2,3,
Patrik Goncalves Rodrigues2,
Hassan Cordash2,
and Douglas C Dean III1,2,4
1Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 2Waisman Center, University of Wisconsin-Madison, Madison, WI, United States, 3Neuroscience Training Program, University of Wisconsin-Madison, Madison, WI, United States, 4Pediatrics, University of Wisconsin-Madison, Madison, WI, United States Keywords: Machine Learning/Artificial Intelligence, Artifacts, Quality Control A three-dimensional convolutional neural network was trained to detect motion artifacts on a volume level for two pediatric diffusion MRI datasets acquired between 1 month and 3 years of age. Accuracies of 95% and 98% were achieved between the two datasets. Additionally, the effects of motion-corrupted volumes on quantitative parameter estimation was examined. Data was processed without quality control and with quality control performed by the neural network. DTI and NODDI metrics were calculated and compared between methods. Significant differences were found for both individual and group results. |
|
4914.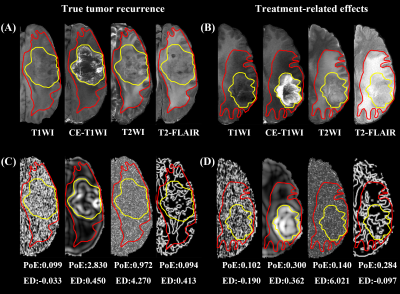 |
148 |
Multimodal MRI radiomics for identifying true tumor recurrence
and treatment-related effects in postoperative glioma patients
Jinfa Ren1,
Dongming Han1,
Xiaoyang Zhai1,
Huijia Yin1,
Ruifang Yan1,
and Kaiyu Wang2
1Department of MR, The First Affiliated Hospital of Xinxiang Medical University, Weihui, China, 2GE Healthcare, MR Research China, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Radiomics Detecting true tumor recurrence and treatment-related effects in glioma after treatment is crucial for patient managements and challenging via conventional MRI for differentiation. Radiomics can be used to access the details in the images in an objective way. We constructed models based on multiple modalities by using radiomics features of the postoperative enhanced and edematous regions to find key features for identifying true tumor recurrence. Features from CE-T1WI and enhanced regions have excellent classification performance, and the model of multimodality with whole regions is the best, which may aid clinicians in developing individualized treatment strategies. |
|
4915.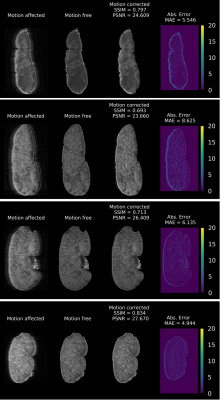 |
149 |
An unsupervised deep learning-based method for in vivo high
resolution Kidney MRI motion correction
Shahrzad Moinian1,2,
Nyoman Kurniawan 1,
Shekhar Chandra 3,
Viktor Vegh1,2,
and David Reutens1,2
1Centre for Advanced Imaging, The University of Queensland, St Lucia, Brisbane, Australia, 2Australian Research Council Training Centre for Innovation in Biomedical Imaging Technology, The University of Queensland, Brisbane, Australia, 3School of Information Technology and Electrical Engineering, The University of Queensland, Brisbane, Australia Keywords: Machine Learning/Artificial Intelligence, Motion Correction A primary challenge for in vivo kidney MRI is the presence of different types of involuntary physiological motion, affecting the diagnostic utility of acquired images due to severe motion artifacts. Existing prospective and retrospective motion correction methods remain ineffective when dealing with complex large amplitude nonrigid motion artifacts. We introduce an unsupervised deep learning-based method for in vivo kidney MRI motion correction. We demonstrate that our deep learning model achieved the average structural similarity index measure (SSIM) of 0.76±0.06 between the reconstructed motion-corrected and ground truth motion-free images, showing an improvement of about 0.33 compared to the corresponding motion-corrupted images. |
|
4916.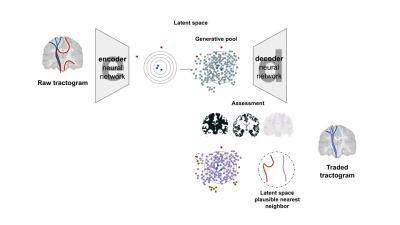 |
150 |
Trading Streamlines in Tractography using Autoencoders (TINTA)
Jon Haitz Legarreta1,
Laurent Petit2,
Pierre-Marc Jodoin1,
and Maxime Descoteaux1
1Computer Science, Université de Sherbrooke, Sherbrooke, QC, Canada, 2Université Bordeaux, Bordeaux, France Keywords: Machine Learning/Artificial Intelligence, Tractography & Fibre Modelling Current tractography methods have a limited ability to accurately reconstruct the long-range brain white matter fiber pathways. Local orientation propagation methods provide tractograms with a non-negligible amount of implausible streamlines. In this work, we propose an artificial intelligence model to recover long-range white matter tracks that are potentially missed in conventional streamline propagation. Our method uses the generative ability of an autoencoder to propose new, plausible streamlines that are subsequently exchanged, according to a given similarity index, with the implausible streamlines in a given tractogram. This allows to potentially improve the reliability of the reconstructed fiber pathways. |
|
4917.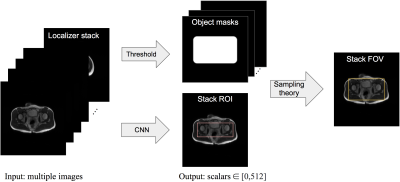 |
151 |
Region of Interest Prediction by Intra-stack Attention Neural
Network
Ke Lei1,
Ali B. Syed2,
Xucheng Zhu3,
John M. Pauly1,
and Shreyas V. Vasanawala2
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States, 3GE Healthcare, Menlo Park, CA, United States Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Region of Interest Manual prescription of the field of view (FOV) by MRI technologists is variable and prolongs the scanning process. Often, the FOV is too large or crops critical anatomy. We propose a deep-learning framework, trained by radiologists’ supervision, for predicting region of interest (ROI) and automating FOV prescription. The proposed ROI prediction model achieves an average IoU of 0.867, significantly better (P<0.05) than two baseline models and not significantly different from a radiologist (P>0.12). The FOV prescribed by the proposed framework achieves an acceptance rate of 92% from an experienced radiologist. |
|
| 4918. | 152 |
Free water imaging parameter estimation by combination of
synthetic q-space learning and conventional fitting: a hybrid
approach
Keigo Yamazaki1,2,
Yoshitaka Masutani3,
Wataru Uchida1,
Koji Kamagata1,
Koh Sasaki4,5,
and Shigeki Aoki1
1Department of Radiorogy, Juntendo University graduate School of Medicine, Tokyo, Japan, 2Department of Radiological Science, Tokyo Metropolitan University, Tokyo, Japan, 3Tohoku University graduate School of Medicine, Miyagi, Japan, 4Graduate School of Infomation Sciences, Hiroshima City University, Hiroshima, Japan, 5Hiroshima Heiwa Clinic, Hiroshima, Japan Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence Free water imaging (FWl), among the diffusion MRI (dMRI) family, is an extended version of the single diffusion tensor model by adding the isotropic diffusion compartment. Generally, FWI parameters have been estimated by fitting of signal model to measured DWI signals. Recently, machine learning techniques have shown promising results also in dMRI parameter inference.In this study, we aimed at development of a hybrid approach for FWI parameter estimation based on synthetic q-space learning (synQSL) and conventional fitting.Our approach was validated by comparison with the conventional fitting method based on quantitative and visual evaluation and computation time. |
|
4919.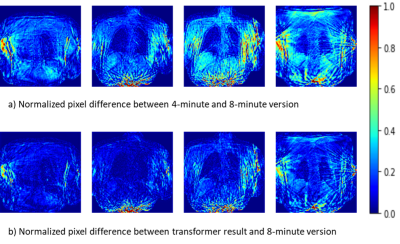 |
153 |
Transformer-based image quality improvement of radial
undersampled lung MRI data from post-COVID-19 patients
Maximilian Zubke1,2,
Robin A Müller1,2,
Marius Wernz1,2,
Filip Klimeš1,2,
Frank Wacker1,2,
and Jens Vogel-Claussen1,2
1Institute of Diagnostic and Interventional Radiology, Hannover Medical School, Hannover, Germany, 2Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH), German Center for Lung Research (DZL), Hannover, Germany Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence Currently, MR-based ventilation imaging relying on radial 3D-stack-of-stars spoiled gradient echo sequence requires a fairly long acquisition time of 8 minutes, which may impact clinical translation. Therefore, a shorter acquisition time is desired. In this study, a novel deep learning approach called transformer was evaluated for image restauration of radial undersampled lung images from 16 post-COVID-19 patients. For each patient, images resulting from 4- and 8 minutes acquisitions were provided. A transformer was trained to translate the 4-minute-version to the corresponding 8-minute-version and led to a significant image quality improvement, demonstrated by three complementary image similarity metrics. |
|
4920.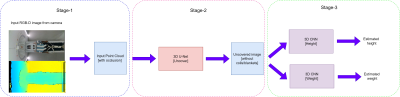 |
154 |
Deep learning based estimation of patient anthropometric data
for intelligent scan planning in MR
Subham Kumar1 and
Harikrishna Rai1
1GE Healthcare, Bengaluru, India Keywords: Machine Learning/Artificial Intelligence, Data Analysis Estimating patient height and weight is used to determine the optimum SAR (Signal Modelling) for the patient. We used a deep learning approach to first generate the point cloud of the person (Data Processing) followed by prediction of the height and weight (Data Analysis). This tackles the problems of heavy occlusion which occurs in the MR imaging scenario in the form of coils/blankets and different positions in which the patient will be placed. We achieved a MAE score of 4.9 cm on the height and 6.2 kg on the weight. This is a promising solution to an important problem. |
|
4921.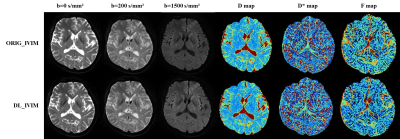 |
155 |
Image Quality and Quantitative Analysis of abbreviated IVIM
Brain MRI With Deep Learning–Based Reconstruction
Qiongge Li1,
Yayan Yin1,
and Jie Lu1
1Xuanwu Hospital Capital Medical University, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Diffusion/other diffusion imaging techniques Deep learning-based reconstruction may improve the image signal to noise ratio without impacting the image contrasts. Intravoxel incoherent motion (IVIM) often require multiple b values with multiple averages that give rise to prolonged scan time. In this work, deep learning reconstruction is used to reduce the overall IVIM scan time. Based on qualitative and quantitative analysis, deep learning reconstruction may significantly improve the results of IVIM and make an abbreviated IVIM feasible. |
|
4922.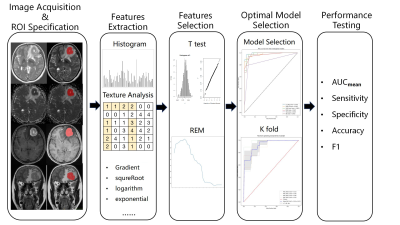 |
156 |
Predicting Double Expression Status in Primary Central Nervous
System Lymphoma Using Multiparametric MRI Based Machine Learning
Li Guo Liu1,
Yue Xin Zhang1,
Nan Zhang1,
Feng Hua Xiao1,
Jing xin Chen1,
and Lin Ma1
1Department of Radiology, The First Medical Center, Chinese PLA General Hospital, Beijing, China Keywords: Machine Learning/Artificial Intelligence, Multimodal In this study, we proposed a promising method to distinguish the double expression lymphoma (DEL) from the non-double expression lymphoma (non-DEL) in primary central nervous system lymphoma (PCNSL) by using multiparametric MRI-based machine learning. The results showed that clinical characteristics and MR imaging features had no significant differences in distinguishing DEL from non-DEL . However, radiomics features could differentiate the two status and the best model in this study was SVMlinear with the combined four sequence group (AUCmean = 0.89±0.04). So multiparametric MRI based machine learning is promising in predicting DEL status in PCNSL. |
|
4923.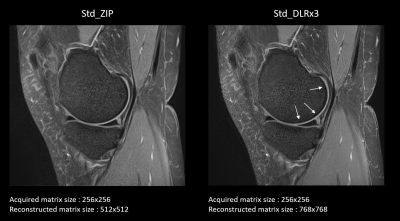 |
157 |
Deep learning-based pipeline to improve sharpness in knee
imaging at both 1.5T and 3T: a clinical evaluation
Valentin H Prevost1,
Shelton Caruthers1,
Khadra Fleury2,
Wissam AlGhuraibawi3,
and Kensuke Shinoda1
1Canon Medical Systems Corporation, Otawara, Japan, 2Canon Medical Systems France, Paris, France, 3Canon Medical Systems USA, Tustin, CA, United States Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence In MRI, edge sharpness is one of the main criteria to allow structure’s delineation and relevant clinical diagnosis. One way to improve the sharpness is to artificially increase the reconstructed matrix size with methods such as zero-padding interpolation (ZIP). In a previous study, we created a deep learning reconstruction (DLR) pipeline, combining ZIP with two CNN’s: the first one trained to reduce image noise, and the second one to reduce ringing artifacts. The goal of this work was to evaluate the clinical impact of this DLR pipeline on pathological knee images performed at 1.5T and 3T, compared to standard reconstructions. |
|
4924.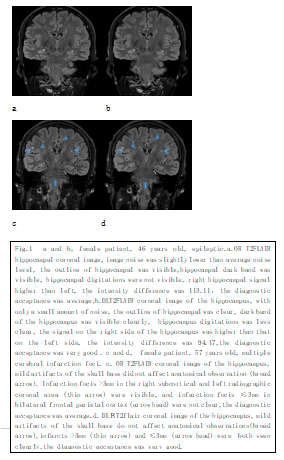 |
158 |
High Resolution Image Quality Improvement of T2 FLAIR Coronal
Hippocampal Imaging by Deep Learning Reconstruction
Yang Jing1,
Li Qiong Ge1,
Wu Tao2,
Qi Zhi Gang1,
Zhao Cheng1,
and Lu Jie1
1Department of Radiology and Nuclear Medicine, xuanwu hospital, Capital Medical University, Beijing, China, 2General Electric Medical System Trade Development (Shanghai) Co., Beijing, China Keywords: Machine Learning/Artificial Intelligence, Brain, Hippocampus, Deep learning reconstruction, Epilepsy Deep learning reconstruction (DL Recon) method can improve the image signal-to-noise ratio and contrast-to-noise ratio without prolonging the scanning time and affecting the signal intensity difference of bilateral hippocampus. It can improve the image quality of hippocampus toe and surrounding small lesions dramatically as in Fig.1. |
|
4925. |
159 |
Association between myosteatosis and impaired glucose
metabolism: A deep learning whole-body MRI population
phenotyping approach
Matthias Jung1,
Marco Reisert2,
Susanne Rospleszcz3,4,
Annette Peters3,4,
Johanna Nattenmüller1,
Christopher L. Schlett1,
Fabian Bamberg1,
and Jakob Weiss1
1Department of Diagnostic and Interventional Radiology, University Medical Center Freiburg, Freiburg, Germany, 2Medical Physics, Department of Diagnostic and Interventional Radiology, University Medical Center Freiburg, Freiburg, Germany, 3Institute of Epidemiology, Helmholtz Zentrum München, Neuherberg, Germany, 4Institute for Medical Information Processing, Biometry and Epidemiology, Ludwig-Maximilians-University München, München, Germany Keywords: Screening, Diabetes Diabetes remains a major challenge for healthcare systems, making screening and early detection desirable. We used deep learning to quantify myosteatosis as 1) skeletal muscle fat fraction (SMFF) and 2) intramuscular adipose tissue (IMAT) normalized for SM mass and assessed their association with impaired glucose metabolism. SMFF had a higher discriminatory capacity for impaired glucose metabolism than IMAT. In multivariable logistic regression adjusted for baseline demographics and cardiometabolic risk factors, only SMFF remained an independent predictor of impaired glucose metabolism. Deep learning-based MR phenotyping enables opportunistic screening of myosteatosis and may identify individuals at high risk for impaired glucose metabolism. |
|
4926.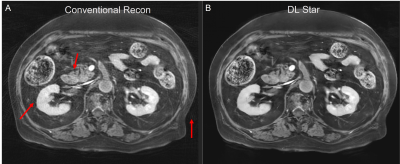 |
160 |
Convolutional Neural Network based Stack-of-Star Imaging with
Noise and Artifacts Removal
Xinzeng Wang1,
Yedaun Lee2,3,
Joonsung Lee4,
Sagar Mandava5,
Ty A. Cashen6,
Xucheng Zhu7,
and Arnaud Guidon8
1GE Healthcare, Houston, TX, United States, 2Department of Radiology, Haeundae Paik Hospital, Busan, Korea, Republic of, 3Inje University College of Medicine, Busan, Korea, Republic of, 4GE Healthcare, Seoul, Korea, Republic of, 5GE Healthcare, Atlanta, GA, United States, 6GE Healthcare, Madison, WI, United States, 7GE Healthcare, Menlo Park, CA, United States, 8GE Healthcare, Boston, MA, United States Keywords: Cancer, Image Reconstruction, Liver, Pancreas Stack of Star acquisition is one of the most frequently used non-Cartesian k-space sampling methods due to its fast speed and robustness to motion. To further reduce the scan time or increase the temporal resolution, Stack of Star is often down-sampled with fewer spokes and advanced sampling patterns, such as golden angle acquisition. However, this makes Stack of Star prone to noise and streak artifacts, limiting the in-plane resolution and degrading diagnostic quality. In this work, we evaluated a deep-learning based stack-of-star method for free-breathing abdominal imaging and it shows improved diagnostic quality. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.