Digital Poster
Data Analysis & Processing I
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
2939.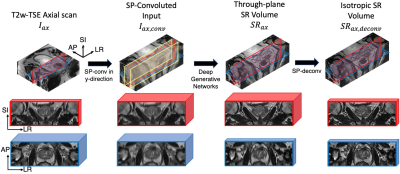 |
61 |
Through-plane Super-resolution of Prostate MRI Using Diffusion
Models
Kai Zhao1,
Jiahao Lin1,
and Kyung Hyun Hyun Sung1
1Department of Radiological Sciences, University of California, Los Angeles, Los Angeles, CA, United States Keywords: Data Analysis, Prostate, super resolution We proposed a generative models-based method for through-plane super-resolution of multi-slice MRI scans. We proposed a novel slice-profile transformation that synthesizes low/high through-plane resolution slices for model training without accessing to isotropic datasets. Expert ready study, visual and quantitative comparisons reveal our method produce superior super-resolution results in terms of sharpness, artifacts, noise level and overall image quality. |
|
2940.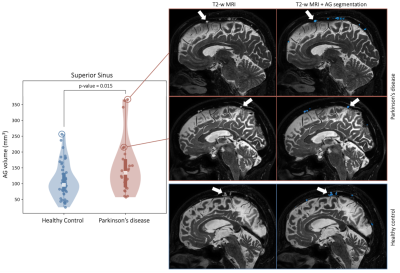 |
62 |
Deep learning of 3D T2-weighted MRI provides support for
arachnoid granulation hypertrophy in patients with Parkinson’s
disease
Melanie Leguizamon1,
Colin McKnight2,
Jarrod Eisma1,
Alex Song1,
Jason Elenberger1,
Daniel Claassen1,
Manus Donahue1,
and Kilian Hett1
1Department of Neurology, Vanderbilt University Medical Center, Nashville, TN, United States, 2Department of Radiology, Vanderbilt University Medical Center, Nashville, TN, United States Keywords: Data Analysis, Neurodegeneration We apply novel deep learning algorithms using non-contrasted T2-weighted MRI to test hypotheses regarding arachnoid granulation (AG) hypertrophy in patients with Parkinson’s disease (PD). Using this method, we identify AGs protruding into the superior sinus lumen, which may serve as a site of CSF egress implicated in the neurofluid clearance system. Results suggest a significant increase in AG volume in the parietal and frontal lobes of PD participants compared to age-matched healthy controls, potentially indicative of reduced neurofluid clearance efficiency secondary to macromolecular aggregation along the CSF circuit. |
|
2941.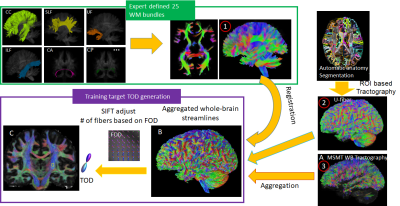 |
63 |
Deep learning improves the estimation of fiber orientation
distribution for tractography in the human
Zifei Liang1,
Patryk Filipiak1,
Steven H Baete1,
Yulin Ge1,
Leslie Ying2,
and Jiangyang Zhang1
1Radiology, NYU Langone health, new york, NY, United States, 2the State University of New York, Buffalo, NY, United States Keywords: Visualization, Brain Connectivity, Diffusion MRI tractography Although diffusion MRI (dMRI) tractography can map brain connectivity non-invasively, accurate tractography in the human brain remains challenging due to inherent and technical limitations. In this study, we demonstrate a deep learning (DL) based approach for improving the estimation of fiber orientation distribution (FOD) from dMRI data. Trained with augmented whole brain tractography results from high-resolution dMRI data, the DL approach outperformed conventional FOD estimation methods in crossing fiber regions with dMRI data at spatial and angular resolutions comparable to routine clinical scans. The approach can potentially shorten the dMRI acquisition necessary for accurate tractography and connectome analysis. |
|
2942.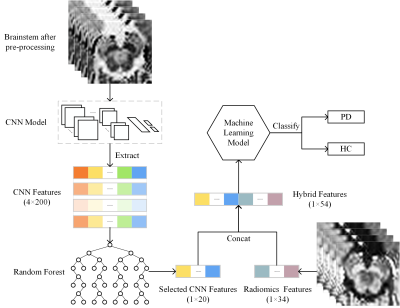 |
64 |
Automated hybrid approach to diagnose Parkinson's disease via
deep learning and radiomics
Hongyi Chen1,
Xueling Liu2,
Yuxin Li1,2,
Puyeh Wu3,
and Daoying Geng1,2
1Academy for Engineering and Technology, Fudan University, Shanghai, China, 2Huashan Hospital, State Key Laboratory of Medical Neurobiology, Fudan University, Shanghai, China, 3GE Healthcare, Beijing, China Keywords: Data Analysis, Parkinson's Disease In this study, we constructed a hybrid machine learning model utilizing CNN and radiomics features based on NM-sensitive setMag images. The hybrid features improved the diagnostic performance in distinguishing PD patients from HC, as demonstrated in the SVM classifier, which demonstrated 95.7% accuracy, 92.9% sensitivity, and 100% specificity. The interpretability of the radiomics approach is better because radiomics features provide more interpretable biomarkers, while the CNN approach extracts deeper features from images. Furthermore, visualizing regions that influence classification decisions via saliency map can also enhance the interpretability of the CNN approach. |
|
2943.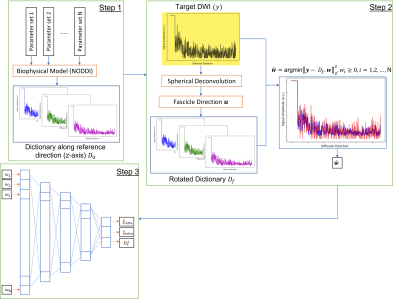 |
65 |
Biophysical model estimation of white matter using machine
learning combined dictionary-based non-negative linear least
square optimization
Hesam Abdolmotalleby1 and
Merry Mani1
1University of Iowa, Iowa City, IA, United States Keywords: Data Processing, Data Processing We estimated biophysical model parameters using a dictionary of diffusion signals. The dictionary is generated based on the set of biophysical model parameters. We then rotated this dictionary along fascicle direction derived from spherical deconvolution and tried to fit it to the diffusion signal using non-negative linear least square optimization to find the associated dictionary weights. These weights went through a deep learning network to find model parameters. Our approach shows reasonable accuracy and reliability for biophysical model estimation. It is also possible to extend our approach to more complex and general biophysical models to achieve higher specificity. |
|
2944.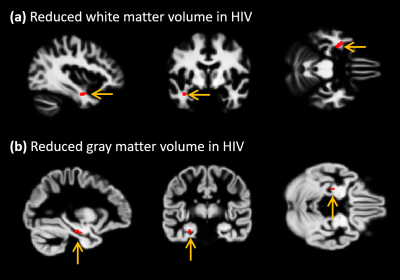 |
66 |
Association of neuroanatomical changes with neuropsychological
changes in Treated Adult HIV-Positive Patients
Ajin Joy1,
Rajakumar Nagarajan1,
Eric Daar2,3,
Jhelum Paul1,
Santosh K Yadav4,
Mario Guerrera2,
Paul M. Macey5,
and M. Albert Thomas1
1Radiological Sciences, University of California, Los Angeles, Los Angeles, CA, United States, 2Division of HIV Medicine, Lundquist Institute at Harbor-UCLA Medical Center, Los Angeles, CA, United States, 3Medicine, University of California, Los Angeles, Los Angeles, CA, United States, 4Department of Radiology and Radiological Science, School of Medicine,, Johns Hopkins University, Baltimore, MD, United States, 5School of Nursing and Brain Research Institute, University of California, Los Angeles, Los Angeles, CA, United States Keywords: Data Processing, Brain This study was designed to use the structural MRI data to characterize volumetric changes in gray matter and white matter, and to determine variations in cortical thickness, in a group of HIV-infected individuals to understand how their brain structural changes are associated with their neuropsychological state compared to a group of healthy individuals with similar social, behavioral backgrounds. Despite the demographic similarities and cART, we observed reduced gray and white matter volumes, as well as altered cortical thickness in HIV-infected participants compared with healthy controls. In addition, the neuroanatomic changes in HIV-infected patients showed statistically significant correlations with memory scores. |
|
2945.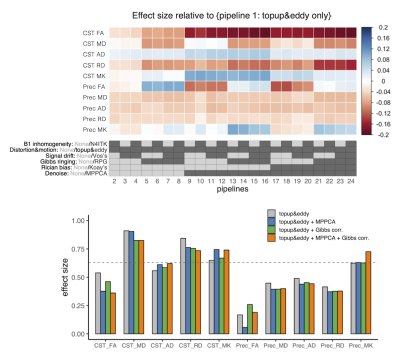 |
67 |
The effects of brain diffusion MRI preprocessing pipelines in a
clinical study of neurodegenerative disease
Kouhei Kamiya1,2,
Sayori Hanashiro3,
Osamu Kano3,
Koji Kamagata2,
Masaaki Hori1,2,
and Shigeki Aoki2
1Department of Radiology, Toho University, Faculty of Medicine, Tokyo, Japan, 2Department of Radiology, Juntendo University, Faculty of Medicine, Tokyo, Japan, 3Department of Neurology, Toho University, Faculty of Medicine, Tokyo, Japan Keywords: Data Processing, Diffusion/other diffusion imaging techniques, Image preprocessing The impacts of different diffusion MRI preprocessing pipelines on the statistical results of clinical studies were investigated in amyotrophic lateral sclerosis. The results illustrate the influences of popular preprocessing tools on the effect size of the disease in DKI metrics. |
|
2946.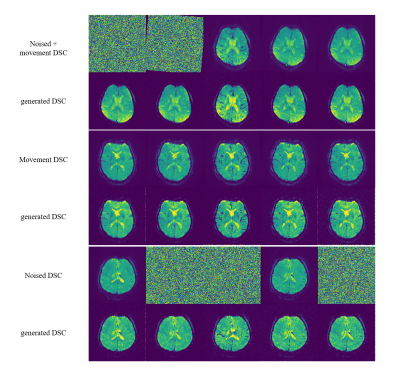 |
68 |
Generative diffusion model for DSC MRI artifact correction
Muhammad Asaduddin1,
Eung Yeop Kim2,
and Sung-Hong Park1
1Bio and Brain Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Department of Radiology, Samsung Medical Center, Sungkyunkwan University College of Medicine, Seoul, Korea, Republic of Keywords: Data Processing, DSC & DCE Perfusion Dynamic susceptibility contrast (DSC) MRI may suffer from artifacts due to long acquisition time. Past methods are limited in their performance and may change the contrast passage timing. In this work, we present a generative diffusion model that can restore signal loss and movement artifacts. We showed the generated DSC MRI images to have proper post-contrast vessel and grey matter structure with accurate contrast agent arrival/washout timing. The brain shape was also accurately generated as shown by the DICE score. This approach could provide a solution to restore a corrupted DSC MRI data while maintaining accurate contrast passage timing. |
|
2947.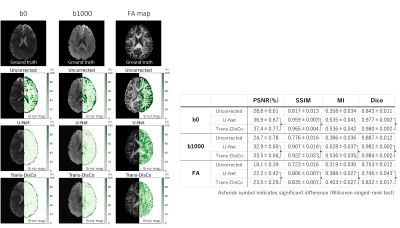 |
69 |
Image distortion correction for diffusion MRI using U-Net and
Transformer
Tsuyoshi Ueyama1,2,3,
Erika Takahashi1,
Naoto Fujita1,
Yuichi Suzuki2,
Hideyuki Iwanaga2,
Osamu Abe4,
and Yasuhiko Terada1
1Graduate School of Science and Technology, University of Tsukuba, Tsukuba, Ibaraki, Japan, 2Radiology center, The University of Tokyo Hospital, Tokyo, Japan, 3School of Medicine, Stanford University, Palo Alto, CA, United States, 4Department of Radiology, The University of Tokyo Hospital, Tokyo, Japan Keywords: Data Processing, Machine Learning/Artificial Intelligence, Diffusion weighted image/Diffusion tensor image Although several end-to-end deep neural networks have been proposed to correct image distortion directly from distorted images, no study has verified the distortion correction performance for high b-values diffusion-weighted image (DWI) and diffusion tensor image (DTI) parameters. For example, the U-Net-based Synb0-DisCo was only validated for distortion correction of b0 images. Here, we used two networks, U-Net and Trans-DisCo, to verify distortion correction performance for DWIs and DTI parameter images. Trans-DisCo is our proposed model that replaces the convolutional neural network in U-Net with Swin Transformer, and we have shown that it outperforms U-Net. |
|
2948.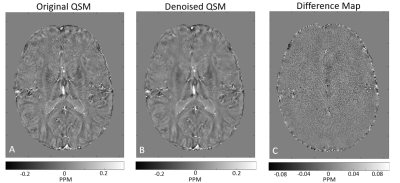 |
70 |
MP-PCA image denoising technique for high resolution
quantitative susceptibility mapping (QSM) of the human brain in
vivo
Liad Doniza1,
Neta Stern2,
Dvir Radunsky2,
Coral Helft3,
Patrick Fuchs4,
Anita Karsa4,
Karin Shmueli4,
and Noam Ben-Eliezer2,3,5
1Department of Electrical Engineering, Tel Aviv University, Tel Aviv, Israel, 2The Department of Biomedical Engineering, Tel-Aviv University, Tel-Aviv, Israel, 3Sagol School of Neuroscience, Tel-Aviv University, Tel-Aviv, Israel, 4Department of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom, 5Center for Advanced Imaging Innovation and Research (CAI2R), New-York University Langone Medical Center, New York, NY, United States Keywords: Data Processing, Susceptibility, Denoising Quantitative susceptibility mapping (QSM) has many clinical applications such as distinguishing between acute and chronic multiple sclerosis (MS) lesions, and probing microbleeds in traumatic brain injury. High scan resolutions improve diagnostic quality and reduce partial volume artifacts albeit at a price of a lower signal-to-noise ratio (SNR). In this study, we introduce a principal component analysis (PCA) denoising algorithm for QSM data, showing the ability to generate QSM maps of the human brain at 0.6x0.6x0.6 mm3 resolution at 3T in vivo. |
|
2949.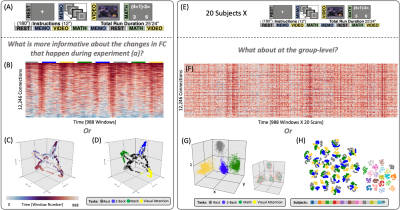 |
71 |
Manifold Learning and Dimensionality Estimation for the Human
Functional Connectome
Javier Gonzalez-Castillo1,
Isabel Fernandez1,
Daniel A Handwerker1,
Ka-Chun Lam2,
Francisco Pereira2,
and Peter A Bandettini1,3
1Section on Functional Imaging Methods, NIMH, Bethesda, MD, United States, 2Machine Learning Team, NIMH, Bethesda, MD, United States, 3FMRI Core Facility, NIMH, Bethesda, MD, United States Keywords: Machine Learning/Artificial Intelligence, fMRI, Manifold Learning, TSNE, UMAP, Laplacian Eigenmaps, Intrinsic Dimension The exploration of time-varying aspects of the human functional connectome (FC) is challenging because the high dimensionality of connectivity matrices precludes direct visual inspection in a meaningful manner. Dimensionality reduction helps circumvent this problem, yet effective application requires a-priori knowledge of the intrinsic dimension of the data and careful selection of algorithmic hyperparameters. Here, we first estimate the intrinsic dimension of FC data. Next, we use data with known cognitive state changes to evaluate the effectiveness of Laplacian Eigenmaps, T-SNE and UMAP to generate informative low dimensional representations of time-varying FC data for explorative and predictive purposes. |
|
2950.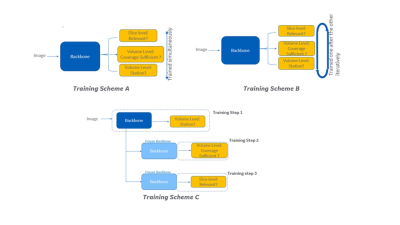 |
72 |
Realizing a robust multi-task training strategy with deep
learning: application in Spine MR image quality assessment
Deepa Anand1,
Dattesh Shanbhag1,
Chitresh Bhushan2,
and Uday Patil3
1GE Healthcare, Bangalore, India, 2GE Healthcare, Niskayuna, NY, United States, 3GE Healthcaer, Bangalore, India Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence Multi-task training is attractive in AI applications (from memory, processing time and potential data reduction), where tasks have commonality in terms of features, but still requires differentiation for individual outputs derived. In this work we present a methodology to implement robust multi-task learning framework considering various strategies (parallel, iterative and sequential). We tested the approach for image quality assessment of spine MRI localizer images. We demonstrate that the sequential training is the most effective, in preserving an accuracy above the acceptable level while allowing for a save in number of model parameters (50%). |
|
2951.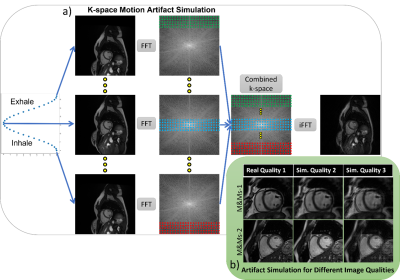 |
73 |
Respiratory Motion Artifact Simulation for DL application in
Cardiac MR Image Quality Assessment
Sina Amirrajab1,
Yasmina Al Khalil1,
Josien Pluim1,
Marcel Breeuwer1,2,
and Cian M. Scannell1
1Biomedical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands, 2MR R&D - Clinical Science, Philips Healthcare, Eindhoven, Netherlands Keywords: Machine Learning/Artificial Intelligence, Artifacts, Respirator Artifact Simulation To tackle data scarcity for training a deep-learning algorithm for cardiac MR image quality assessment, we develop a k-space method for simulating respiratory motion artifacts with different levels of severity on artifact-free publicly available cardiac MRI data. The benefit of such simulated data is investigated, demonstrating the usefulness of training a feature extractor with the simulated artifacts for image quality classification. Our proposed method achieved the test accuracy of 0.625 and Cohen's Kappa of 0.473 (n=120 images), ranking third in task one for the CMRxMotion challenge of MICCAI 2022. |
|
2952.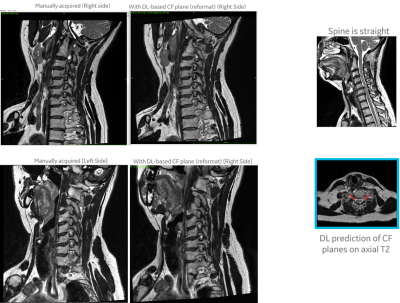 |
74 |
Deep Learning based prediction of the planes for automated
planning of MRI imaging of cervical neural foramina and lumbar
pars interarticularis
Chitresh Bhushan1,
Dattesh D Shanbhag2,
Uday Patil2,
Trevor Kolupar3,
and Maggie Fung3
1AI and Medical Imaging, GE Research, Niskayuna, NY, United States, 2GE Healthcare, Bangalore, India, 3GE Healthcare, Waukesha, WI, United States Keywords: Machine Learning/Artificial Intelligence, Visualization, Scan Planning, Spine, Reformatting We present a generalized DL-based intelligent slice placement framework for planes of cervical neural foramina (CF) and lumbar pars interarticularis (PI) for spine MRI. CF and PI scan improves assessment of foraminal stenosis and lumbar spondylolysis respectively, but requires highly skilled operator for accurate prescription. Our approach enables automatic patient-specific scan plane prescription from routine axial T2W images. In our test, it achieves mean error of <0.7 mm and <0.2 degrees and demonstrates similar/better contiguous anatomical visualization to manual scans on retrospective reformatting of 3D data. These results indicate that our approach is accurate and suitable for clinical usage. |
|
2953.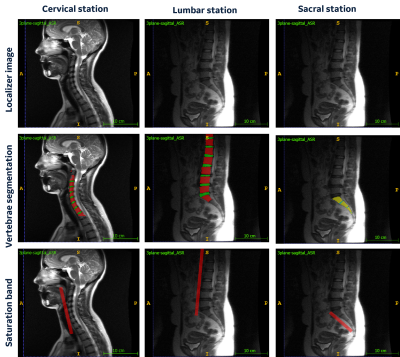 |
75 |
Deep learning-based saturation band placement in spine localizer
images
Ashish Saxena1,
Chitresh Bhushan2,
Soumya Ghose2,
Patil Uday1,
Kameswari Padmanabhan3,
Sanjay NT3,
and Dattesh Shanbhag3
1GE Research, Bengaluru, India, 2GE Research, Niskayuna, NY, United States, 3GE Healthcare, Bengaluru, India Keywords: Machine Learning/Artificial Intelligence, Visualization, workflow Saturation band placement is important for obtaining good quality MRI images in presence of structures which can generate artifacts such as cardiac regions or large pulsating blood vessels. In anatomy such as spine, saturation-band placement can be time consuming since technologist has to ensure that it doesn’t overlap with vertebrae regions. In this study, we demonstrated an automated deep-learning method to accurately place the saturation band on 2D three-plane localizer images with no intervention from the technologist. |
|
2954.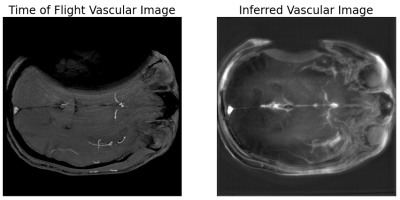 |
76 |
Within Patient Contrast Adjustment Through a Self-Consistent
Deep Learning Model when Imaging Near Metal: An Example in
Angiography
Kevin M. Koch1,2 and
Andrew S. Nencka1,2
1Radiology, Medical College of Wisconsin, Milwaukee, WI, United States, 2Center for Imaging Research, Medical College of Wisconsin, Milwaukee, WI, United States Keywords: Machine Learning/Artificial Intelligence, Vessels, Metal Artifact Multi-spectral imaging around metallic implants allows for a unique application of subject-specific deep learning applications because both high-resolution standard acquisitions and multi-spectral acquisitions are performed with significant regions of artifact-free overlap. A deep neural network can be trained in that region of artifact-free overlap within a single patient to yield a contrast transform on the multi-spectral images and achieve similar contrast to traditional acquisitions in the region of those acquisitions obscured by metal artifacts. In this proof of concept, a preliminary example of synthesizing time of flight-like contrast from a multi-spectral acquisition with vascular flow voids was shown. |
|
2955.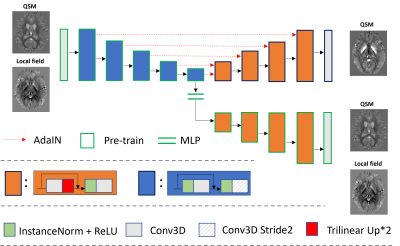 |
77 |
Real-time style transfer for quantitative susceptibility mapping
using unsupervised learning
Zhuang Xiong1,
Yang Gao2,
and Hongfu Sun1
1the University of Queensland, Brisbane, Australia, 2Central South University, Changsha, China Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, unsuperivised learning Existing supervised deep learning methods for quantitative susceptibility mapping (QSM) could lead to degraded results when applied to phase images acquired with different scan parameters, such as image resolution and acquisition orientation. This work proposes a novel unsupervised learning method incorporating style transfer and deep image prior to enabling the reconstruction of susceptibility maps from local field maps acquired with any scan parameters. To speed up the inference, a pre-training strategy is also proposed, reducing reconstruction time from minutes to seconds. |
|
2956.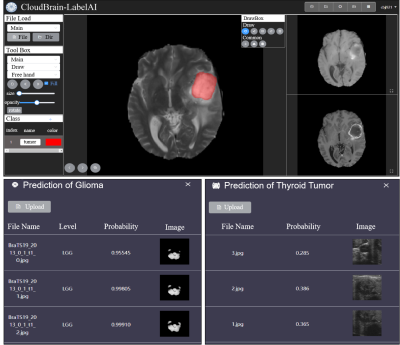 |
78 |
CloudBrain-LabelAI: An Online Intelligent Medical Imaging
Annotation and Training Platform
Bangjun Chen1,
Jian Wang1,
Yirong Zhou1,
Yu Hu1,
Shuxian Niu1,
Biao Qu2,
Di Guo3,
Jingjing Xu1,
Jiyang Dong1,
and Xiaobo Qu1
1Biomedical Intelligent Cloud R&D Center, Department of Electronic Science, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2Department of Instrumental and Electrical Engineering, Xiamen University, Xiamen, China, 3School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China Keywords: Machine Learning/Artificial Intelligence, Segmentation Deep learning and cloud computing technologies have shown remarkable performance in medical imaging research. Using deep learning for clinical research requires a programming foundation, while the annotation of image data is a highly specialized and time-consuming process. In this work, we develop a high-performance online medical image annotation and training platform (CloudBrain-LabelAI). It provides medical image researchers with an efficient image annotation platform for the rapid construction of datasets for deep learning. Meanwhile, it provides codeless image segmentation training and prediction based on cloud computing, which greatly reduces the threshold of medical image segmentation efforts and simplifies the overall workflow. |
|
2957.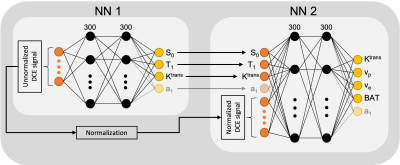 |
79 |
Self-contained comprehensive quantification of dynamic
contrast-enhanced MRI using physics+kinetics-based network
learning (PKNet)
Soudabeh Kargar1,
Ouri Cohen1,
Sungmin Woo2,
Hebert Alberto Vargas2,
and Ricardo Otazo1
1Medical Physics, MSKCC, New York, NY, United States, 2Radiology, MSKCC, New York, NY, United States Keywords: Machine Learning/Artificial Intelligence, DSC & DCE Perfusion, Deep Learning This work develops a deep learning technique that is trained according to physics and kinetics for self-contained comprehensive quantification of dynamic contrast-enhanced MRI. In addition to perfusion parameters, patient-specific parameters that affect the quantification are estimated, including bolus arrival time, T1, steady state magnetization, and AIF. The DCE-MRI quantification network was tested on a patient with cervical cancer and demonstrated high concordance between two scans separated by 24 hours. Physics plus kinetics informed network learning (PKNet) enabled the quantification of multiple parameters which has the potential to increase reproducibility of quantitative DCE-MRI, a long-desired goal. |
|
2958.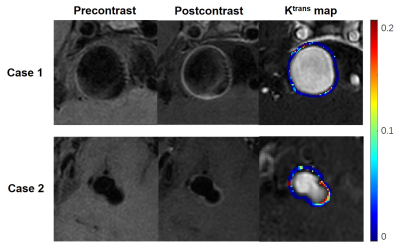 |
80 |
Predicting the Risk of Non-Saccular Aneurysms Based on the
Dynamic Contrast-Enhanced Magnetic Resonance Imaging
Yan Li1,
Ziming Xu1,
Linggen Dong2,
Yajie Wang1,
Peng Liu2,
Ming Lv2,
and Huijun Chen1
1Center for Biomedical Imaging Research, Tsinghua University, Beijing, China, 2Department of Neurosurgery, Beijing Tiantan Hospital, Beijing Neurosurgical Institute, Capital Medical University, Beijing, China Keywords: Data Analysis, DSC & DCE Perfusion Assessment of intracranial aneurysm rupture risk is clinically critical. Recent studies have shown that the aneurysm wall permeability (Ktrans) and wall enhancement index are independent risk factors for predicting saccular aneurysm rupture. However, this conclusion has not been confirmed in non-saccular aneurysms. Our study suggested that Ktrans was significantly associated with the aneurysm size and PHASES score in non-saccular aneurysms. But Ktrans did not associate with the wall enhancement index in non-saccular aneurysms. Therefore, Ktrans can provide an independent quantitative indicator for assessing the risk of rupture of in non-saccular intracranialaneurysms. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.