Digital Poster
Spectroscopy, MT, CEST
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Computer # | |||
|---|---|---|---|
3429.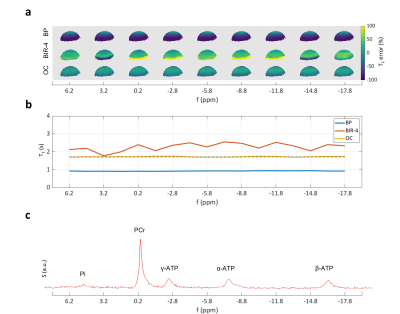 |
21 |
Optimal control RF pulses for dual-angle T1 measurements in 31P
magnetization transfer spectroscopy
Clemens Diwoky1,
Christina Graf2,
Armin Rund3,
and Rudolf Stollberger2
1Institute of Molecular Biosciences, University of Graz, Graz, Austria, 2Institute of Biomedical Imaging, Graz University of Technology, Graz, Austria, 3Institute for Mathematics and Scientific Computing, University of Graz, Graz, Austria Keywords: Pulse Sequence Design, CEST & MT Herein, we show for the first time the application of optimal control (OC) to the design of RF pulses for dual-angle T1 measurements as needed for 31P magnetization transfer spectroscopy. Phantom measurements highlight that OC RF pulses are able to produce precise T1 maps within the relevant coil's sensitivity range. The resulting enlarged spin ensemble leads to high SNR and excellent T1 prediction if used in non-selective spectroscopic application. In addition, OC pulses were optimized for operation at a large bandwidth suitable for 31P spectroscopy (± 15 ppm), which is often a problem when adiabatic BIR-4 pulses are used. |
|
3430.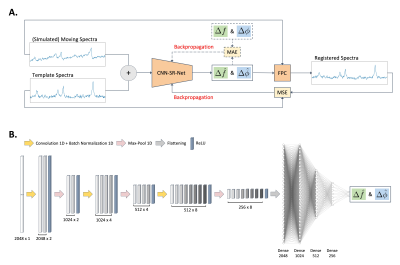 |
22 |
Magnetic Resonance Spectroscopy Spectral Registration with
Unsupervised Deep Learning
David Jing Ma1,
Yanting Yang1,
Natalia Harguindeguy1,
Ye Tian1,
Scott A. Small2,3,4,
Feng Liu2,5,
Douglas L. Rothman6,
and Jia Guo2,7
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Department of Psychiatry, Columbia University, New York, NY, United States, 3Department of Neurology, Columbia University, New York, NY, United States, 4Taub Institute Research on Alzheimer's Disease and the Aging Brain, Columbia University, New York, NY, United States, 5New York State Psychiatric Institute, Columbia University, New York, NY, United States, 6Radiology and Biomedical Imaging of Biomedical Engineering, Yale University, New Haven, CT, United States, 7Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States Keywords: Data Processing, Machine Learning/Artificial Intelligence A deep learning-based registration method has been a successful image processing tool adopted in medical image registration but there is a lack of learning-based registration tools for spectral registration protocols. A novel CNN-based unsupervised deep learning spectral registration model was developed and trained on a simulation dataset. The model was then further evaluated on a simulated test set with more extreme conditions and on an in vivo dataset and was compared performances to published frequency-and-phase correction models. An unsupervised deep learning-based spectral registration approach was found to demonstrate state-of-the-art performance in frequency-and-phase correction. |
|
3431.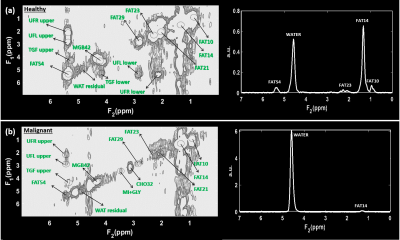 |
23 |
Association of Ki-67 With Metabolite and Lipid Levels in High
Grade Breast Cancer Patients based on Correlated MR
Spectroscopic Imaging and Biopsy
Ajin Joy1,
Andres Saucedo1,
Melissa Joines1,
Stephanie Lee-Felker1,
Manoj K Sarma1,
James Sayre1,
Maggie Dinome2,
and M. Albert Thomas1
1Radiological Sciences, University of California, Los Angeles, Los Angeles, CA, United States, 2Surgery, University of California, Los Angeles, Los Angeles, CA, United States Keywords: Data Analysis, Breast Five-dimensional (5D) echo-planar correlated spectroscopic imaging (EP-COSI) combines 2 spectral and 3 spatial dimensions to record two dimensional (2D) correlated spectroscopy (COSY) spectra in multiple regions in multiple slices. In this study, multiple 2D COSY spectra were recorded from breast cancer patients by 5D EP-COSI within practical scan time durations using non-uniform undersampling of one spectral and two spatial dimensions, and compressed sensing-based reconstruction. Different metabolite and lipid ratios were quantified and its association with Ki-67 metric was studied. Findings of this study showed statistically significant association of metabolite and lipid levels with Ki-67 measures in breast cancer patients. |
|
3432.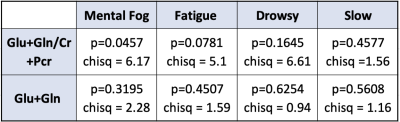 |
24 |
Glutamate Predicts Post-Concussion Symptoms in Collegiate
Athletes
Annelise Lemaire1,2,
Hui Jun Liao1,
Inga Koerte3,
David Howell4,
Alexander Lin1,5,
and Katherine Breedlove1,5
1Brigham and Women’s Hospital, Boston, MA, United States, 2University of Michigan, Ann Arbor, MI, United States, 3Ludwig-Maximilians-Universitat, Munich, Germany, 4University of Colorado-Anschutz, Aurora, CO, United States, 5Harvard Medical School, Boston, MA, United States Keywords: Data Analysis, Contrast Mechanisms, Traumatic Brain Injury A population of collegiate athletes, along with matched age and gender-matched student-athlete controls, were evaluated post-concussion, for neurometabolic levels in the PCG by magnetic resonance spectroscopy (MRS) and Post-Concussion Symptom Scale (PCSS) following injury. Evaluations were conducted 72 hours post-diagnosis. For the purpose of this work, correlations between total Glutamate (Glu+Gln), total Glutamate to total Creatine ratios, and symptoms on the PCS scale associated with cognitive fatigue - such as mental fog, drowsiness, fatigue, and feeling slowed - were explored. |
|
3433.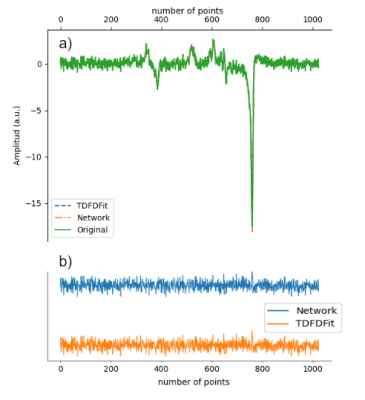 |
25 |
MRS Quantification using Deep Learning Frameworks: an Accuracy
and Efficiency Study
Federico Turco1 and
Johannes Slotboom2
1Support Center for Advanced Neuroimaging (SCAN), University of Bern, Bern, Switzerland, 2Support Center for Advanced Neuroimaging (SCAN), Institute of Diagnostic and Interventional Neuroradiology, Bern, Switzerland Keywords: Data Processing, Spectroscopy, Deep learning frameworks, Deep learning, Optimization We implemented a model fitting algorithm for magnetic resonance spectroscopy quantification using Pytorch. This network fit the spectra by minimizing the error between the spectrum modeled by the newtwork and the desired target. The implementation can fit multiple spectras in parallel using Pytorch GPU acceleration. We compared the results with a parallel version of TDFDFit and found it to be up to 12 time faster fitting 2048 spectras in 50 seconds. |
|
3434.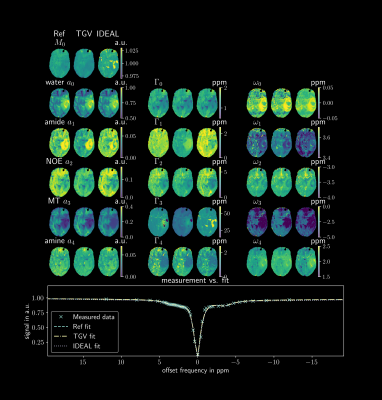 |
26 |
Performance of Multipool Lorentzian fitting for CEST-MRI using
joint spatial regularization
Markus Huemer1,
Oliver Maier1,
Moritz Simon Fabian2,
Manuel Schmidt2,
Arnd Dörfler2,
Kristian Bredies3,
Moritz Zaiss2,4,
and Rudolf Stollberger1,5
1Institute of Biomedical Imaging, Graz University of Technology, Graz, Austria, 2Institute of Neuroradiology, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), University Hospital Erlangen, Erlangen, Germany, 3Institute of Mathematics and Scientific Computing, University of Graz, Graz, Austria, 4High-Field Magnetic Resonance Center, Max-Planck Institute for Biological Cybernetics, Tübingen, Germany, 5BioTechMed Graz, Graz, Austria Keywords: Data Analysis, CEST & MT This work investigates an improved quantification algorithm for CEST spectra using a joint Frobenius TGV regularization. The performance is assessed with simulations, phantom and in-vivo measurement with respect to systematic errors and robustness. For comparison, a pixel-wise fit using the Levenberg-Marquardt algorithm and the IDEAL algorithm were implemented. The $$$TGV_j$$$ algorithm shows the best performance in the main parameter fitting stability and parameter SNR. |
|
3435.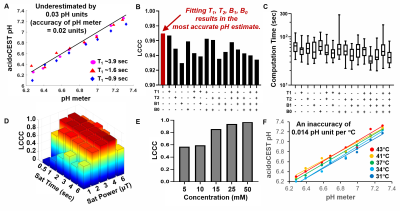 |
27 |
Improved Bloch fitting and machine learning methods that analyze
acidoCEST MRI
Tianzhe Li1,2,
Julio Cardenas-Rodriguez3,
and Marty David Pagel4
1Cancer Systems Imaging, UT MD Anderson Cancer Center, Houston, TX, United States, 2Medical Physics Program, UT Health, Houston, TX, United States, 3Data Translators LLC, Oro Valley, AZ, United States, 4Cancer Systems Imaging, MD Anderson Cancer Center, Houston, TX, United States Keywords: Data Analysis, CEST & MT, pH imaging AcidoCEST MRI can measure the extracellular pH of the tumor microenvironment. We have further refined our “Bloch fitting” method, and shown that this analysis method can accurately and precisely measure pH without additional MRI information, with an accuracy of 0.03 pH units. In addition, we have developed a machine learning method that can classify pH as > 7.0 or < 6.5 pH units (PPV=0.94, NPV=096), and a machine learning regression method that can estimate pH with a mean absolute error of 0.031 pH units. |
|
3436.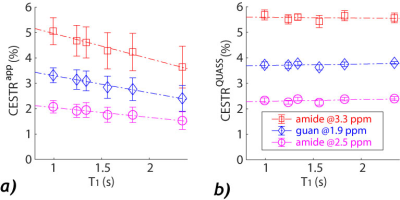 |
28 |
Demonstration of multi-pool quantitative CEST MRI with
quasi-steady-state reconstruction – Insight into T1-independent
CEST quantification
Lauren Gao1 and
Phillip Sun1
1Emory University, Atlanta, GA, United States Keywords: Data Analysis, CEST & MT CEST quantification is challenging because the measurement depends on the scan protocols. Also, T1 normalization is not straightforward under non-equilibrium conditions. Using multi-pool simulations and phantom experiments, our study evaluated quasi-steady-state(QUASS) algorithm-boosted CEST quantification. The 3-pool CEST simulation showed significant T1 dependencies due to complex interactions among Ts, TR, and T1. Such dependencies were corrected with QUASS reconstruction. In addition, a multi-T1 phantom was used to evaluate the quantification. Whereas the apparent CEST MRI showed significant dependence on Ts, TR, and T1, accurate CEST quantification was demonstrated with the spinlock-model-based fitting of QUASS CEST MRI. |
|
3437.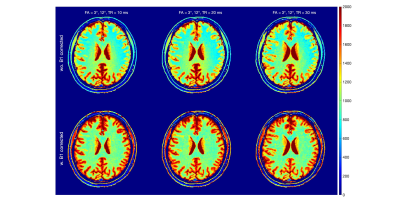 |
29 |
The Influence of the MT effect on the Accuracy of T1
Quantification by the Dual Flip Angle Method
Zhen Hu1,
Dan Zhu2,3,
and Qin Qin2,3
1Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Kirby Center, Kennedy Krieger Institute, Baltimore, MD, United States Keywords: Signal Modeling, Brain, Relaxometry The dual flip angle (DFA) method is widely used for brain T1 quantification. In addition to the well-known RF field effect, this study investigated the overlooked magnetization transfer (MT) effect for the accuracy of in-vivo T1 mapping. Based on the two-pool Bloch simulation, the DFA derived T1 estimation revealed FA and TR dependence, which was confirmed by the in-vivo brain T1 mapping using the same set of sequence parameters. Longer TR (30 ms) reduced the variation between FA pairs and were closer to the literature values, but at the cost of longer acquisition time. |
|
3438.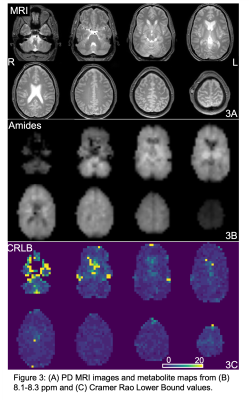 |
30 |
Downfield Proton MRSI with whole-brain coverage at 3T
İpek Özdemir1,
Sandeep Ganji2,
Joseph Gillen1,3,
Michal Považan4,
and Peter B Barker1,3
1Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University, Baltimore, MD, United States, 2Philips Healthcare, Best, Netherlands, 3F.M. Kennedy Krieger Institute, Baltimore, MD, United States, 4Danish Research Centre for MR, Copenhagen, Denmark Keywords: Data Acquisition, Spectroscopy, Downfield MRSI A 3D downfield-MRSI sequence with whole brain coverage, 22-minute scan time, and 0.7 cm3 nominal spatial resolution has been developed at 3T. The sequence was tested in 5 normal volunteers; LCModel analysis showed CRLB average values of 3-4% for protein amide resonances in 3 selected gray and white matter regions. |
|
3439.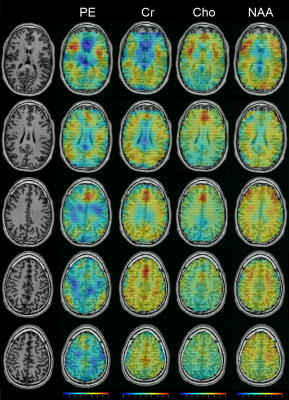 |
31 |
In vivo Whole-brain Phosphoethanolamine Mapping using SLOW-EPSI
at 7T
Guodong Weng1,2,
Piotr Radojewski1,2,
Federico Turco1,2,
and Johannes Slotboom1,2
1Support Center for Advanced Neuroimaging (SCAN), University of Bern, Bern, Switzerland, 2Translational Imaging Center, sitem-insel, Bern, Switzerland Keywords: Visualization, Spectroscopy, Spectral editing In vivo detection of phosphoethanolamine (PE) using spectral editing has recently been shown to be feasible. This study shows whole-brain PE maps using SLOW-EPSI in a healthy subject with a nominal resolution of 4.3*4.3*10 mm. The result shows that the PE level is high in the cerebral cortex and low in the white matter. |
|
| 3440. | 32 |
EigenMRS: A computationally cheap data-driven approach to MR
spectroscopic imaging denoising
Amirmohammad Shamaei1,2,
Jana Starcukova1,
and Zenon Starcuk Jr1
1Institute of Scientific Instruments of the Czech Academy of Sciences Research institute in Brno, Brno, Czech Republic, 2Department of Biomedical Engineering, Brno University of Technology, Brno, Czech Republic Keywords: Data Processing, Spectroscopy, Singular value decomposition, MR spectroscopic imaging, Denoising
The utility of MR spectroscopic imaging (MRSI) can be
limited by a low signal-to-noise ratio (SNR) in practice.
Averaging multiple coherent repetitions increases the SNR,
but at the cost of time-consuming acquisition. Several
computationally expensive approaches based on low-rank
matrix approximation for denoising MRSI data have been
proposed, which do not take advantage of previously acquired
spectra. |
|
3441.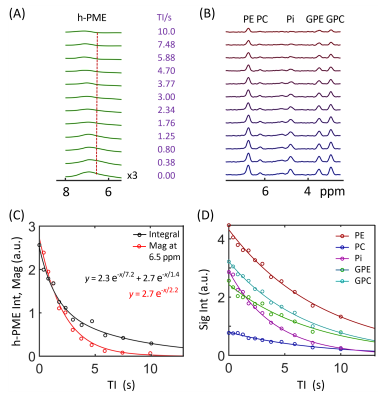 |
33 |
Revealing hidden phosphomonoester signals under
phosphoethanolamine and phosphocholine resonances: A brain 31P
MRS study at 7T
Jimin Ren1,2
1Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX, United States, 2Department of Radiology, University of Texas Southwestern Medical Center, Dallas, TX, United States Keywords: Data Analysis, Spectroscopy, brain, phosphocholine, phosphoethanolamine, phosphomonoester It has been a common practice to quantify brain phosphomonoester (PME) 31P signals using a two-component model composed of phosphoethanolamine (PE) and phosphocholine (PC). This study demonstrates spectral evidence of the presence of a hidden broad PME (h-PME) signal underneath PE and PC resonances, characterized by a short T1 and potentially contributed by RBC 2,3-DPG in brain blood vessels, though other sources of signal contribution cannot be fully ruled out. The results have implication in using PE and PC as biomarkers of altered phospholipid metabolism in brain pathologies. |
|
3442.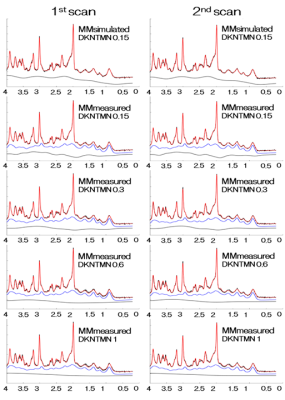 |
34 |
Analysis of short-TE STEAM data at 7T: Effects of a
macromolecular basis set and baseline parameters on GABA,
glutamate and their ratio.
Tomohisa Okada1,
Hideto Kuribayashi2,
Yuta Urushibata2,
Thai Akasaka1,
Ravi Teja Seethamraju3,
Sinyeob Ahn3,
and Tadashi Isa1
1Human Brain Research Center, Kyoto University, Kyoto, Japan, 2Siemens Healthcare, Japan, Tokyo, Japan, 3Siemens Medical Solutions, Malvern, PA, United States Keywords: Data Analysis, Spectroscopy The effect of baseline flexibility LCModel parameter DKNTMN (0.15, 0.3, 0.6 and 1) on a 7T short-TE STEAM MRS measurement of GABA, glutamate and excitatory-inhibitory ratio (EIR) was investigated using both simulated and measured macromolecular basis sets. Mean (SD) of GABA/tCr was highest, 0.23 (0.02), using simulated MM basis. Using measured MM basis, the ratios decreased from 0.18 (0.04) to 0.12 (0.03) by the increase of DKNTMN values. The GABA/tCr ratio and EIR of a former multi-center study was 0.19 and 8.2 after T2 decay correction. Analysis using DKNTMN of 0.3 conformed best and considered to be the choice. |
|
3443.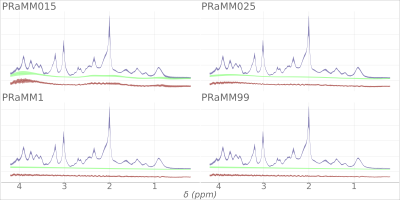 |
35 |
Parametrized macromolecule models in the brain 1H MRS
quantification: Effect of the baseline spline on quantification
uncertainty and precision
Andrea Dell'Orco1,2,
Layla Tabea Riemann2,
Stephen L. R. Ellison3,
Bernd Ittermann2,
Anna Tietze1,
Michael Scheel1,
and Ariane Fillmer2
1Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt- Universität zu Berlin, Department of Neuroradiology, Berlin, Germany, Berlin, Germany, 2Physikalisch-Technische Bundesanstalt (PTB), Braunschweig und Berlin, Germany., Berlin, Germany, 3LGC Limited Registered, Teddington, Middlesex, United Kingdom Keywords: Data Analysis, Spectroscopy, macromolecules, single-voxel 1HMRS Parametric macromolecule (MM) models can improve the quantification of brain 1H-MRS. We quantified a publicly available repeated-acquisition dataset with LCModel and applied a parametric MM model in conjunction with four different settings for spline node distance and compared the quantification results. Coefficients of variation, average Cramér-Rao lower bound, repeatability, and reproducibility were estimated. A moderate effect of the baseline spline was shown. |
|
3444.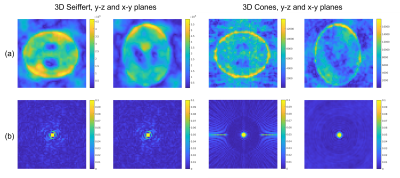 |
36 |
3D Seiffert spirals for efficient k-space sampling in 23Na-MRI:
initial phantom simulations
Samuel Rot1,2,
Matthew Clemence3,
Bhavana S Solanky1,4,
and Claudia AM Gandini Wheeler-Kingshott1,5,6
1NMR Research Unit, Queen Square MS Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, United Kingdom, 2Department of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom, 3Philips Healthcare, Best, Netherlands, 4Quantitative Imaging Group, Department of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom, 5Department of Brain & Behavioural Sciences, University of Pavia, Pavia, Italy, 6Brain Connectivity Centre Research Department, IRCCS Mondino Foundation, Pavia, Italy Keywords: New Trajectories & Spatial Encoding Methods, Non-Proton Modern sodium MRI (23Na-MRI) uses efficient, non-Cartesian k-space sampling strategies to acquire 3D volumes in clinically feasible scan times. 3D Seiffert spirals are a novel k-space sampling scheme with improved efficiency to existing methods. 23Na-MRI, and particularly temporally resolved functional 23Na-MRI, could be an ideal application for Seiffert spirals. We present initial, in silico simulations of 3D Seiffert spiral k-space sampling for a highly undersampled 1.8 second functional 23Na-MRI protocol. Seiffert spirals generate images of improved quality and SNR in direct comparison to 3D-cones. Essential further work will involve compressed sensing reconstruction, further trajectory optimisation and in vivo imaging. |
|
3445.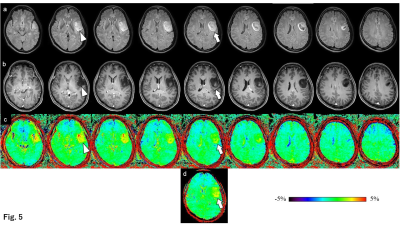 |
37 |
Grading and assessment of intra-tumor heterogeneity of gliomas
using 3D CEST imaging with compressed sensing and sensitivity
encoding
Tatsuhiro Wada1,2,
Osamu Togao3,
Chiaki Tokunaga1,
Kazufumi Kikuchi4,
Koji Yamashita5,
Masami Yoneyama6,
Masahiro Oga1,
Koji Kobayashi1,
Toyoyuki Kato1,
Kousei Ishigami4,
and Hidetake Yabuuchi7
1Division of Radiology, Department of Medical Technology, Kyushu university hospital, Fukuoka, Japan, 2Department of Health Sciences, Graduate school of Medical Sciences, Kyushu University, Fukuoka, Japan, 3Department of Molecular Imaging & Diagnosis, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan, 4Department of Clinical Radiology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan, 5Department of Radiology Informatics & Network, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan, 6Philips Japan, Tokyo, Japan, 7Department of Health Sciences, Faculty of Medical Sciences, Kyushu University, Fukuoka, Japan Keywords: Tumors, CEST & MT Glioma grading using chemical exchange saturation transfer (CEST) imaging is often performed in a single cross-section. However, CEST imaging of multiple cross-sections is desirable for intra-tumor heterogeneity. Compressed sensing and sensitivity encoding (CS-SENSE) was applied to CEST imaging to obtain multi-slice CEST imaging in a clinically appropriate scan time. The diagnostic performance of three-dimensional (3D) CEST imaging was comparable to that of a two-dimensional CEST imaging. The evaluation of the entire tumors by multi-slice CEST imaging was important in the gliomas' grading because the signal intensities differed among the tumor slices. |
|
3446.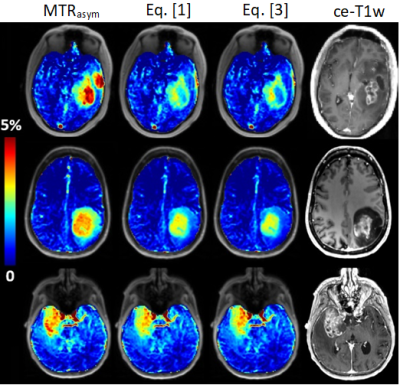 |
38 |
Theoretical justification for fluid-suppressed APTw imaging
based on spillover correction principles
Maria Sedykh1,
Stefano Casagranda2,
Patrick Liebig3,
Christos Papageorgakis2,
Laura Mancini4,5,
Sotirios Bisdas4,5,
Manuel Schmidt1,
Arnd Doerfler1,
and Moritz Zaiss1
1Institut of Neuroradiology, University Clinic Erlangen, Friedrich-Alexander Universität Erlangen-Nürnberg, Erlangen, Germany, 2Department of R&D Advanced Applications, Olea Medical, La Ciotat, France, 3Siemens Healthcare GmbH, Erlangen, Germany, 4Lysholm Department of Neuroradiology, University College of London Hospitals NHS Foundation Trust, London, United Kingdom, 5Institute of Neurology UCL, London, United Kingdom Keywords: Tumors, CEST & MT Fluid suppression has an inestimable value in improving the readability of Amide Proton Transfer weighted imaging in the neuro-oncological field, by removing contamination effects in the fluid compartments. With this work we wanted to justify the derivation of this metric and to show different clinical examples. Our new fluid-suppressed APTw metric for 3T MR imaging, based on the principles of Spillover Correction for CEST imaging, can be then derived through the Bloch McConnell Equations. |
|
3447.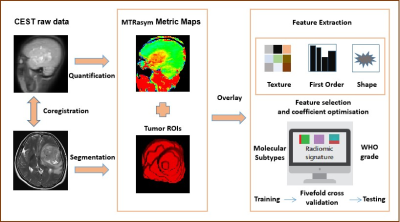 |
39 |
Amide proton transfer weighted (APTw) imaging-based radiomics in
the molecular subtypes prediction and grading of adult-type
diffuse gliomas
Minghao Wu1,
Tongling Jiang2,
Cong Xie3,
Xianchang Zhang4,
Yi Zhang5,
and Yaou Liu1
1Department of Radiology, Beijing Tiantan Hospital, Capital Medical University, Beijing, China, beijing, China, 2Department of Biomedical Engineering, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, Zhejiang, China, Hangzhou, China, 3Beijing Tiantan Hospital, Capital Medical University, Beijing, China, beijing, China, 4MR Collaboration, Siemens Healthineers Ltd., Beijing, China, beijing, China, 5College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, Zhejiang, China, Hangzhou, China Keywords: Radiomics, Brain, amide proton transfer weighted (APTw) imaging The fifth edition of the 2021 WHO Classification of Tumors of the Central Nervous System (WHO CNS5) has introduced significant changes in classifying glioma subtypes based on molecular profiles. This study investigated the feasibility of amide proton transfer weighted (APTw)-based radiomics for predicting molecular subtypes and WHO grade of adult-type diffuse gliomas. APTw-based radiomics achieved satisfactory performance in distinguishing three molecular subtypes and WHO grade (High (Ⅳ grade)/Low (Ⅱ-Ⅲ grade)). This application may be an effective and promising quantitative approach for better noninvasive characterization and classification of gliomas. |
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.