Oral Session
Novel Advances in Segmentation
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

08:15 |
0804.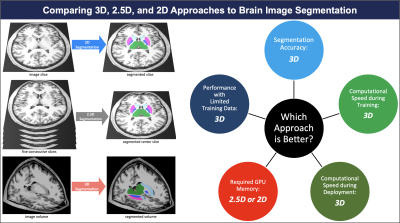 |
Comparing 3D, 2.5D, and 2D Approaches to Brain MRI Segmentation
Arman Avesta1,
Sajid Hossain1,
MingDe Lin2,
Mariam Aboian2,
Harlan M Krumholz3,
and Sanjay Aneja4 1Therapeutic Radiology, Yale Unviersity, New Haven, CT, United States, 2Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States, 3Division of Cardiovascular Medicine, Yale Unviersity, New Haven, CT, United States, 4Therapeutic Radiology, Yale University, New Haven, CT, United States Keywords: Segmentation, Segmentation We compared 3D, 2.5D, and 2D approaches to brain MRI auto-segmentation and concluded that the 3D approach is more accurate, achieves better performance when training data is limited, and is faster to train and deploy. Our results hold across various deep-learning architectures, including capsule networks, UNets, and nnUNets. The only downside of 3D approach is that it requires 20 times more computational memory compared to 2.5D or 2D approaches. Because 3D capsule networks only need twice the computational memory that 2.5D or 2D UNets and nnUNets need, we suggest using 3D capsule networks in settings where computational memory is limited. |
08:23 |
0805.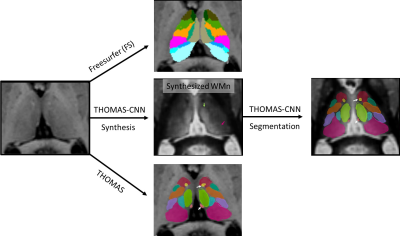 |
Thalamic segmentation methods in the characterization of
Alzheimer's disease
Brendan Williams1,
Dan Nguyen2,
and Manojkumar Saranathan2
1University of Reading, Reading, United Kingdom, 2University of Massachusetts Chan Medical School, Worcester, MA, United States Keywords: Segmentation, Segmentation We present a systematic comparison of three state of the art thalamic segmentation methods for T1 MRI. Segmentation performance against Krauth-Morel atlas was quantified on 100 young healthy subjects and thalamic atrophy as a function of Alzheimer's disease status was characterized on 540 older subjects. |
| 08:31 |
0806. |
Artifact-robust vascular segmentation for 3D phase-contrast MR
angiography using a deep learning approach
Daiki Tamada1,
Thekla H Oechtering1,2,
Eisuke Takai3,
and Scott B Reeder1,4,5,6,7
1Radiology, University of Wisconsin-Madison, Madison, WI, United States, 2Department of Radiology, Universität zu Lübeck, Lübeck, Germany, 3MIRAI Technology Institute, Shiseido Co., Ltd., Tokyo, Japan, 4Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 5Biomedical Engineering, University of Wisconsin-Madison, Madison, WI, United States, 6Medicine, University of Wisconsin-Madison, Madison, WI, United States, 7Emergency, University of Wisconsin-Madison, Madison, WI, United States Keywords: Segmentation, Segmentation, phase contrast MRA We developed a segmentation algorithm for PC-MRA using a deep-learning approach, with the goal of achieving artifact-robust segmentation for PC-MRA. To simulate flow-related artifacts of MRA, Gaussian noise and phase error were added to the k-space domain of the datasets. LadderNet consists of two consecutive U-nets with skip connections, and has been adopted as a training network for vessel segmentation. Retrospective studies demonstrated superior accuracy and precision of the proposed method over a conventional level set segmentation method. |
| 08:39 |
0807.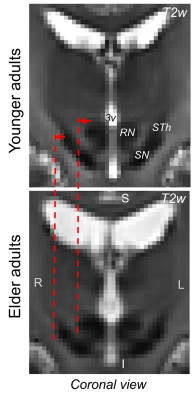 |
Towards a probabilistic structural atlas of brainstem nuclei in
elderly humans using in-vivo 7 Tesla multi-contrast MRI
Subhranil Koley1,
Kavita Singh1,
Maria G. Garcia-Gomar1,2,
and Marta Bianciardi1,3
1Brainstem Imaging Laboratory, Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Boston, MA, United States, 2Escuela Nacional de Estudios Superiores Unidad Juriquilla, Universidad Nacional Autónoma de México, Queretaro, Mexico, 3Division of Sleep Medicine, Harvard University, Boston, MA, United States Keywords: Segmentation, Aging, Brainstem atlas Brainstem nuclei are involved in several vital functions; impairment in their structure and function is manifested in several clinical conditions of elderly humans, including sleep/arousal/movement/vestibular/anxiety disorders, chronic pain and altered autonomic functions. Brainstem evaluation in health and disease is currently limited by the difficulty of localizing these regions in conventional MRI. We developed an in-vivo probabilistic atlas of 31 brainstem nuclei in elderly humans by the use of multi-contrast 7 Tesla MRI in 15 elderly subjects and of an existing brainstem nuclei atlas in younger adults. This atlas can be applied to conventional MRI and aid brainstem investigation in aging. |
| 08:47 |
0808.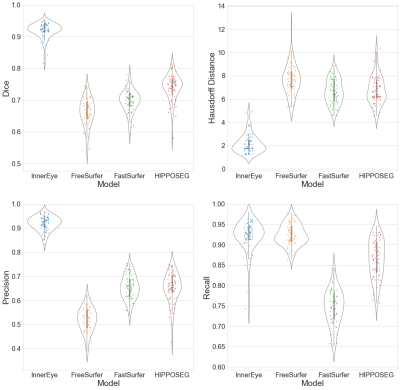 |
InnerEye as a Tool for Accurate Hippocampal Segmentation
Anna Schroder1,
James Moggridge2,3,
Jiaming Wu1,4,
Hamza A. Salhab2,3,
Sjoerd Vos5,
Melissa Bristow6,
Fernando Pérez-García6,
Javier Alvarez-Valle6,
Tarek A. Yousry2,3,
John S. Thornton2,3,
Frederik Barkhof1,3,4,7,
Matthew Grech-Sollars1,2,
and Daniel C. Alexander1
1Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 2Lysholm Department of Neuroradiology, National Hospital for Neurology and Neurosurgery, University College London Hospitals NHS Foundation Trust, London, United Kingdom, 3Department of Brain Repair and Rehabilitation, UCL Institute of Neurology, University College London, London, United Kingdom, 4Department of Medical Physics & Biomedical Engineering, University College London, London, United Kingdom, 5Centre for Microscopy, Characterisation & Analysis, University of Western Australia, Perth, Australia, 6Health Futures, Microsoft Research Cambridge, Cambridge, United Kingdom, 7Department of Radiology and Nuclear Medicine, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam UMC, Amsterdam, Netherlands Keywords: Segmentation, Brain, Hippocampus Accurate hippocampal segmentation tools are critical for monitoring neurodegenerative disease progression on MRI and assessing the impact of interventional treatment. Here we present the InnerEye hippocampal segmentation model and evaluate this new model against three standard segmentation tools in an Alzheimer’s disease dataset. We found InnerEye performed best for Dice score, precision and Hausdorff distance. InnerEye performs consistently well across the different cognitive diagnoses, while performance for other methods decreased with cognitive decline. |
| 08:55 |
0809.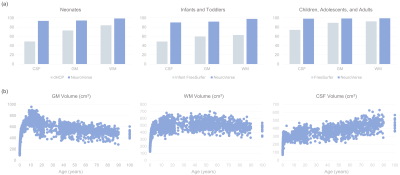 |
NeuroVerse: Neuroimage Processing Tools Across a Century of Life
Sahar Ahmad1,
Xiaoyang Chen1,
Wenjiao Lyu1,
Jinjian Wu1,
Yifan Li1,
Yicheng Zou1,
Kim-Han Thung1,
Siyuan Liu1,
and Pew-Thian Yap1
1Department of Radiology and Biomedical Research Imaging Center (BRIC), The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States Keywords: Segmentation, Brain, Surface reconstruction; Lifespan analysis The availability of large-scale multi-site neuroimaging data present unprecedented opportunities for revealing the brain's macroscopic and microscopic organization across the lifespan. Existing neurodevelopmental studies lack consensus, owing to variable preprocessing methods that result in inconsistent brain features. Existing neuroimage processing tools typically cater to specific life periods, limiting their applicability to data covering the entire human lifespan. Here, we present robust automatic tools for accurate and consistent processing of neuroimaging data covering a century of life. |
| 09:03 |
0810.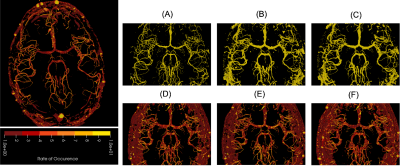 |
Exploiting the inter-rater disagreement to improve probabilistic
segmentation
Soumick Chatterjee1,2,3,
Franziska Gaidzik4,
Alessandro Sciarra3,5,
Hendrik Mattern3,
Gabor Janiga4,
Oliver Speck3,6,7,
Andreas Nürnberger1,2,7,
and Sahani Pathiraja8
1Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4Laboratory of Fluid Dynamics and Technical Flows, Otto von Guericke University Magdeburg, Magdeburg, Germany, 5MedDigit, Department of Neurology, Medical Faculty, University Hospital Magdeburg, Magdeburg, Germany, 6German Center for Neurodegenerative Disease, Magdeburg, Germany, 7Center for Behavioral Brain Sciences, Magdeburg, Germany, 8School of Mathematics & Statistics, University of New South Wales, Sydney, Australia Keywords: Segmentation, Brain Disagreements among the experts while segmenting a certain region can be observed for complex segmentation tasks. Deep learning based solution Probabilistic UNet is one of the possible solutions that can learn from a given set of labels for each individual input image and then can produce multiple segmentations for each. But, this does not incorporate the knowledge about the segmentation distribution explicitly. This research extends the idea by incorporating the distribution of the plausible labels as a loss term. The proposed method could reduce the GED by 47% and 63% for multiple sclerosis and vessel segmentation tasks. |
| 09:11 |
0811.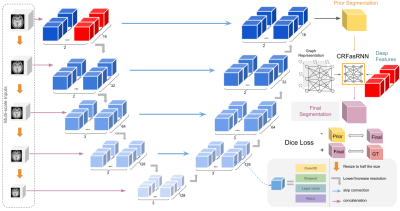 |
EVAC: Multi-scale V-Net with Deep Feature Conditional Random
Field Layers for Brain Extraction
Jong Sung Park1,
Shreyas Fadnavis2,
and Eleftherios Garyfallidis1
1Intelligent Systems Engineering / Neuroscience, Indiana University, Bloomington, Bloomington, IN, United States, 2Harvard University, Cambridge, MA, United States Keywords: Segmentation, Brain Brain Extraction is a complicated semantic segmentation task. While Deep Learning methods are popularly used, they are heavily biased towards the training dataset. To reduce this dependency, we present EVAC (Enhanced V-net like Architecture with Conditional Random Fields), a novel Deep Learning model for Brain Extraction. Using V-net as a skeleton, we propose three improvements: multi-scale inputs, modified CRF layer and regularizing Dice Loss. Results show that these changes not only increase accuracy but also the efficiency of the model as well. Compared to the state-of-the-art methods, our model achieves high and stable accuracy across datasets. |
| 09:19 |
0812.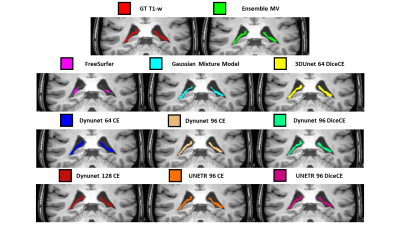 |
The ensemble of optimized Deep Learning Neural Networks improves
the estimate of the Choroid Plexus Volume: application to
Multiple Sclerosis
Valentina Visani1,
Valerio Natale2,
Annalisa Colombi3,
Agnese Tamanti3,
Alessandra Bertoldo1,
Corina Marjin3,
Francesca Benedetta Pizzini2,
Massimiliano Calabrese3,
and Marco Castellaro1
1Department of Information Engineering, University of Padova, Padova, Italy, 2University Hospital of Verona, Verona, Italy, 3Department of Neurosciences, Biomedicine and Movement Sciences, University of Verona, Verona, Italy Keywords: Segmentation, Machine Learning/Artificial Intelligence, Choroid Plexus The Choroid Plexus (ChP) is a brain vascular tissue involved in regulatory processes. ChP Volume (ChPV) modifications are related to neurodegenerative disorders, consequently, it was suggested the use of ChPV as biomarker. This work proposes a method for the automatic segmentation of ChP based on Deep-Learning Neural-Networks (DNNs) hyperparameters optimization. Ninety-Six hyperparameters and architectures combinations were trained on T1-w MRI in MONAI, first selection was made on bias and variance and best DNNs were ensembled by major voting. Ensemble model outperforms single DNNs and freely available software (FreeSurfer, Gaussian Mixture Model), highlighting the ensembles DNNs exploitability to automatically estimate ChPV. |
| 09:27 |
0813.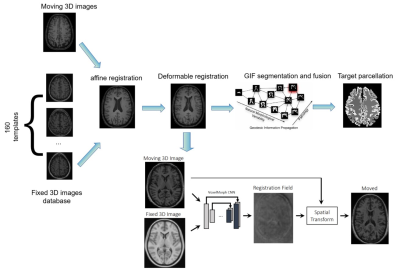 |
GIF_boost : A Generalisable Hybrid Brain Tissue Segmentation
with DeepLearning
JIAMING WU1,
Giuseppe Pontillo1,2,
Zoe Mendelsohn1,2,3,
Yipeng Hu1,
Frederik Barkhof1,4,
and Ferran Prados1,2,5
1Centre for Medical Image Computing (CMIC), University College London, London, United Kingdom, 2Neuroradiological Academic Unit, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom, 3Department of Radiology, Charité School of Medicine and University Hospital Berlin, Berlin, Germany, 4Radiology & Nuclear Medicine, VU University Medical Center, Amsterdam, Netherlands, 5e-Health Centre, Universitat Oberta de Catalunya, Barcelona, Spain Keywords: Segmentation, Segmentation Image segmentation and parcellation can provide quantitative assessment of the brain and can guide diagnosis and treatment decision-making. Geodesic Information Flow (GIF) is a freely available brain tissue segmentation and parcellation MRI-based tool using a classical label fusion approach. In this work, we introduce GIF_boost, a hybrid solution that takes advantages of deep learning to accelerate the bottleneck step of the template library registration. We compared GIF_boost with the original version of GIF and FreeSurfer (a state-of-the-art method). GIF_boost performed parcellation minimum 16 times faster. Parcellations had a similar Dice coefficient and Hausdorff distance and an improved volumetric quantification. |
| 09:35 |
0814.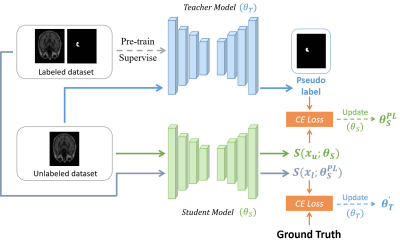 |
Segmentation of Brain Structures MR Images via Semi-Supervised
Learning
Rui Li1,
Ziming Xu1,
Runyu Yang1,
Jiaqi Dou1,
Tianyi Yan2,
and Huijun Chen1
1Center for Biomedical Imaging Research, School of Medicine, Tsinghua University, Beijing, China, 2School of Life Science, Beijing Institute of Technology, Beijing, China Keywords: Segmentation, Brain The precise segmentation of brain structures is of great importance in quantitatively analyzing brain medical resonance images. In recent years, more and more experts attempt to apply semi-supervised learning to medical image segmentation tasks, since it could make use of the rich unlabeled data. Based on this, we modified a segmentation model based on semi-supervised learning for automatically segmenting brain structures. We tested model on two hippocampus public datasets and results show that our model has considerable segmentation performance compared with that of model trained in a supervised manner, which illustrates the effectiveness and potential of our model. |
| 09:43 |
0815.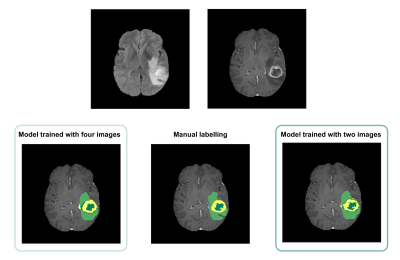 |
Automated Brain Tumour Segmentation in Glioblastoma: Can similar
performance be achieved using a shorter imaging protocol?
Catarina Passarinho1,
Oscar Lally2,
Patrícia Figueiredo1,
and Rita G. Nunes1
1Institute for Systems and Robotics - Lisboa and Department of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal, 2King's College London, London, United Kingdom Keywords: Segmentation, Brain The same deep learning model was trained for automated segmentation of glioblastoma tumour regions using either four or two MRI modalities. The performance of the model trained with only two images was found to be comparable to that of the longer protocol, suggesting that the excluded images and the consequent longer training time did not contribute significantly to the accuracy of the model. These findings strongly imply that this training approach may be beneficial for clinical applications, as it would result in reduced costs due to shorter scanner times, lower computational requirements and increased patient throughput, without compromising segmentation accuracy. |
| 09:51 |
0816.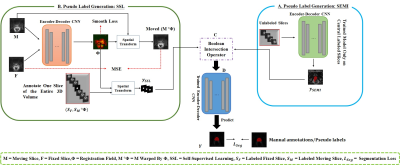 |
Sparse Annotation Deep Learning for Prostate Segmentation of
Volumetric Magnetic Resonance Images
Yousuf Babiker M. Osman1,2,
Cheng Li1,3,
Weijian Huang1,2,4,
Nazik Elsayed1,2,5,
Zhenzhen Xue1,3,
Hairong Zheng1,
and Shanshan Wang1,3,4
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University of Chinese Academy of Sciences, Beijing, China, 3Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application, Guangzhou, China, 4Peng Cheng Laboratory, Shenzhen, China, 5Faculty of Mathematical and Computer Sciences, University of Gezira, Wad Madani, Sudan Keywords: Segmentation, Prostate Deep neural networks (DNNs) have achieved unprecedented performances in various medical image segmentation tasks. Nevertheless, DNN training requires a large amount of densely labeled data, which are labor-intensive and time-consuming to obtain. Here, we address the task of segmenting volumetric MR images using extremely sparse annotations, for which only the central slices are labeled manually. In our framework, two independent sets of pseudo labels are generated for unlabeled slices using self-supervised and semi-supervised learning methods. Boolean operation is adopted to achieve robust pseudo labels. Our approach can be very important in clinical applications to reduce manual effort on dataset construction. |
09:59 |
0817.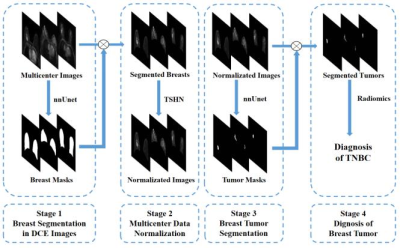 |
Automatic Segmentation and Diagnosis of Breast Cancer in
Multicenter Data based on Deep Learning and Tissue-specific
Histogram Normalization
Yansong Bai1,
Rencheng Zheng1,
Weibo Chen2,
Chao You3,
Chengyan Wang4,
and He Wang1
1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Philips Healthcare, Shanghai, China, 3Shanghai Cancer Center, Fudan University, Shanghai, China, 4Human Phenome Institute, Fudan University, Shanghai, China Keywords: Segmentation, Breast This study presented an intelligent diagnosis system to segment breast tumors in dynamic contrast-enhanced (DCE) images from multicenter dataset and determine the risk of triple-negative breast cancer (TNBC) with a four-step model: a) breast segmentation with no-new Unet (nnUnet); b) multicenter data normalization using Tissue-specific Histogram Normalization (TSHN); c) tumor segmentation with nnUnet model and d) automatic diagnosis of breast cancer with radiomics analysis based on segmented masks. The proposed model exhibited a superior performance in segmentation and diagnosis of breast cancer in multicenter data.
|
| 10:07 |
0818.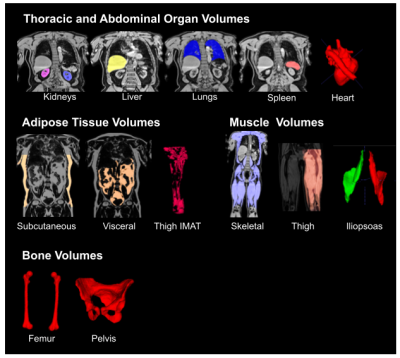 |
Automated Processing and Segmentation of Abdominal Structures
Using a Hybrid Attention-Convolutional Neural Network Model
Nicolas Basty1,
Ramprakash Srinivasan2,
Marjola Thanaj1,
Elena P Sorokin2,
Madeleine Cule2,
E Louise Thomas1,
Jimmy D Bell1,
and Brandon Whitcher1
1University of Westminster, London, United Kingdom, 2Calico Life Sciences LLC, South San Francisco, CA, United States Keywords: Data Analysis, Body, Deep learning, Dixon Automated image processing and organ segmentation are critical to the quantitative analysis of population-scale imaging studies. We have implemented an end-to-end pipeline for neck-to-knee Dixon MRI data based on the UK Biobank abdominal protocol. Bias-field correction, blending across series boundaries, and fat-water swap correction are performed in the preprocessing steps. A hybrid attention-convolutional neural network model segments multiple abdominal organs, major bones, along with adipose and muscle tissue. The application of neural network models, to both swap detection and segmentation, produces a computationally-efficient pipeline that scales to accommodate tens of thousands of datasets. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.