Oral Session
Data Analysis & Processing
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

08:15 |
0069.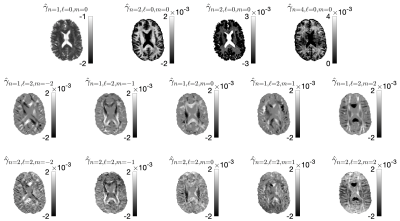 |
What if every voxel was measured with a different diffusion protocol?
Santiago Coelho1, Gregory Lemberskiy1, Ante Zhu2, Nastaren Abad2, Thomas K. F. Foo2, Els Fieremans1, and Dmitry S. Novikov1
1Center for Advanced Imaging Innovation and Research (CAI2R), Department of Radiology, New York University School of Medicine, New York, NY, United States, 2GE Research, Niskayuna, NY, United States Keywords: Data Processing, Diffusion/other diffusion imaging techniques A naive answer to this question is to loop over all voxels re-do the training and apply machine learning estimators of your favorite model. This is grossly computationally inefficient. We propose a matrix pseudoinversion-based method that can estimate nonlinear biophysical model parameters from a large set of voxels with independent acquisition protocols in a few minutes. Our framework can be tailored to any convolution-based model. Furthermore, the protocols are not required to have shells and there are no limits to the protocol differences among voxels. This method is readily extendable for simultaneously varying diffusion times, B-tensor shapes, TE, etc. |
| 08:23 | 0070.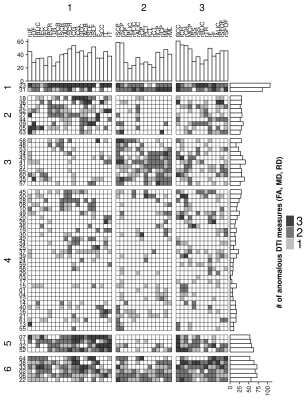 |
Empirical normative models to identify patterns of anomalous white matter in autism spectrum disorder
Nagesh Adluru1, Douglas Dean III1, Molly Prigge2, Jace King2, Nicholas Lange3, Erin Bigler4, Brandon Zielinski5, Janet Lainhart1, and Andrew Alexander1
1University of Wisconsin, Madison, WI, United States, 2University of Utah, Salt Lake City, UT, United States, 3Harvard Medical School, Cambridge, MA, United States, 4Brigham Young University, Provo, UT, United States, 5University of Florida, Gainesville, FL, United States Keywords: Data Analysis, Brain Normative modeling offers a promising approach for better characterization of brain differences in heterogeneous populations. In this study, empirical normative modeling was applied to characterize white matter regional heterogeneity of DTI measurements in adolescents and young adults with more cognitively-and-verbally able autism spectrum disorder (ASD) in comparison to an age matched cohort of typically developing controls. The results of this study demonstrated that the individual differences in DTI measurements in many JHU white matter regions are extremely heterogeneous across the ASD cohort. Hierarchical clustering analysis revealed three groups of white matter regions with similarity patterns across six subgroups within ASD. |
08:31 |
0071.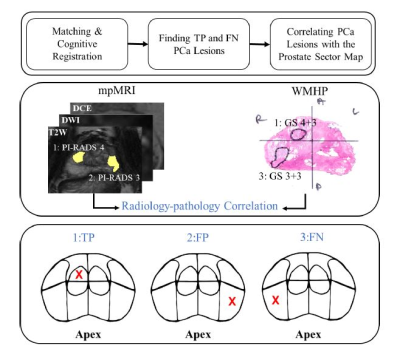 |
Analyzing the Spatially-Resolved Performance of mpMRI for Detection of Prostate Cancer using a Prostate Sector Map
Fatemeh Zabihollahy1, Steven S Raman1, Pornphan Wibulpolprasert2, Sitaram Vangala3, Holden H. Wu1, Aichi Chien1, Albert Thomas1, Robert E. Reiter4, and Kyunghyun Sung1 1Department of Radiological Sciences, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, United States, 2Department of Diagnostic and Therapeutic Radiology, Ramathibodi Hospital, Bangkok, Thailand, 3Department of Medicine Statistics Core, University of California, Los Angeles, Los Angeles, CA, United States, 4Department of Urology, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, United States Keywords: Data Analysis, Multimodal Multiparametric MRI (mpMRI) has shown a marked impact on prostate cancer (PCa) diagnosis, but the understanding of the spatial distribution of tumors and the correlation between tumor location and mpMRI performance is still limited. The purpose of this study was to determine the performance of mpMRI for PCa detection and visualize the results on a prostate sector map described by Prostate Imaging Reporting and Data System (PI-RADS) v2.1. Also, this study aimed to investigate the association between mpMRI performance and tumor location in the prostate. |
| 08:39 | 0072. | Intravoxel incoherent motion DWI with different mathematical models in predicting rectal adenoma with and without canceration
Jia Yuping1 and Dou Weiqiang2
1Lixia District, Jinan city, The First Affiliated Hospital of Shandong First Medical University & Shandong Provincial Qianfoshan, Jinan, China, 2MR Research, GE Healthcare, Beijing, China Keywords: Data Analysis, Cancer This study aimed to use intravoxel incoherent motion (IVIM) DWI with different mathematical models to predict rectal adenomas with and without canceration. The parameters of different IVIM-DWI models, including apparent diffusion coefficient (ADC) from mono-exponential model, true diffusion coefficient, pseudo-diffusion coefficient and perfusion fraction from bi-exponential model, and the distributed diffusion coefficient and water molecular diffusion heterogeneity index from stretched-exponential model were compared between 31 adenoma and 29 adenoma with canceration. We found that mono-exponential derived ADC can easily predict rectal adenomas with canceration, and bi-exponential model has a better combination of sensitivity and specificity for diagnosing rectal adenoma canceration. |
| 08:47 | 0073.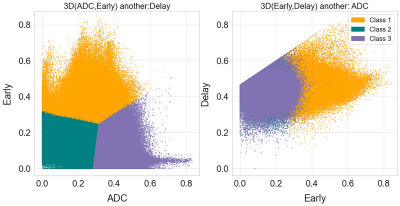 |
Measurement of tumor heterogeneity with habitats in breast cancer and its application in molecular subtype discrimination
Run Xu1, Guanwu Li1, Dan YU2, Yongming Dai2, and Xinyue Liang2
1Department of Radiology, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China, 2Central Research Institute, United Imaging Healthcare, Shanghai, China Keywords: Data Analysis, Cancer In this work, multiparametric MRI was applied to reconstruct spatial habitats and validate the association with the molecular subtypes of breast cancer. By combining perfusion and diffusion characteristics, three habitats were constructed and assigned: hypervascular habitat, hypovascular cellular habitat, and nonviable habitat. In triple-negative breast cancer (TNBC), the volume fraction is lower for hypervascular habitat and higher for nonviable habitat, with respect to non-TNBC. |
| 08:55 | 0074.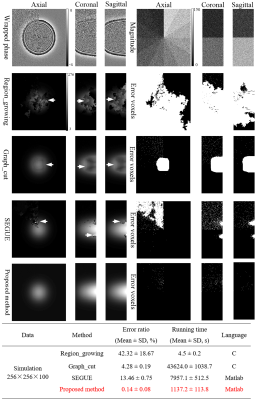 |
A Novel Accelerated 3D Phase-Unwrapping Method Based on Subdivided Arrays and Polynomial Modeling with Application: Dixon Water-Fat Separation
Junying Cheng1, Qian Zheng2, Man Xu1, Liang Liu1, Yan Cui1, Biaoshui Liu3, Yong Zhang1, Yanqiu Feng4, and Jingliang Cheng1
1Department of MRI, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 2College of Software Engineering, Zhengzhou University of Light Industry, Zhengzhou, China, 3Department of Radiation Oncology, Sun Yat-sen University CancerCenter, Guangzhou, China, 4School of Biomedical Engineering and Guangdong Provincial Key Laboratory of Medical Image Processing, Southern Medical University, Guangzhou, China Keywords: Data Processing, Data Processing, phase imaging, phase unwrapping, Dixon technique In this work, a novel robust and accelerated phase-unwrapping method is presented. The proposed method firstly introduces an artificial volume compartmentalization to break down the large-scale unwrapping problems, and then uses the phase partition method to cluster the phase into blocks to be paralleled unwrapped first, and residual-voxel to be unwrapped later. The simulated and in vivo datasets experiments have demonstrated that the proposed method allows for a reduction of the unwrapping problem size, a speed-up for handling large datasets, and obtains the accurate phase results under different SNRs and phase-change levels. |
| 09:03 | 0075. |
Reducing the impact of disrupted brain regions in Diffusion Tensor Imaging with inpainting
Zihao Tang1,2, Xinyi Wang2, Mariano Cabezas1, Lihaowen Zhu2, Dongnan Liu1,2, Michael Barnett1,3, Weidong Cai2, and Chenyu Wang1,3
1Brain and Mind Centre, University of Sydney, Camperdown, Australia, 2School of Computer Science, University of Sydney, Camperdown, Australia, 3Sydney Neuroimaging Analysis Centre, Camperdown, Australia Keywords: Data Analysis, Diffusion Tensor Imaging Diffusion Weighted Imaging (DWI) can be disrupted due to acquisition constraints or imaging artifacts, which can lead to unreliable scalar metrics calculated and valuable scans discarded as a result. To reduce the impact of disrupted brain regions in DTI, we adapted a deep learning DTI inpainting network to reconstruct the disrupted ROIs. We evaluated the performance of the method by calculating the Fractional Anisotropy errors according to each individual brain region. Experimental results show that the inpainting method can reconstruct the relevant clinical imaging information by mitigating the Fractional Anisotropy differences overall and in individual disrupted brain regions. |
| 09:11 | 0076.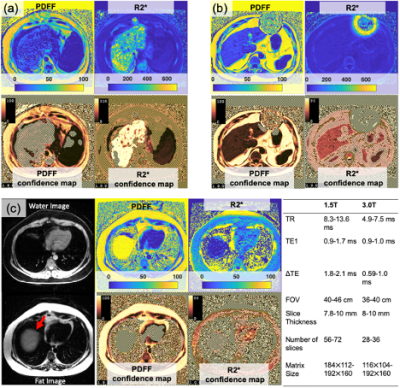 |
Confidence Maps for Reliable Proton Density Fat-Fraction and R2* Estimation
Daiki Tamada1, Rianne A Van der Heijden1, Diego Hernando1,2,3,4, and Scott B Reeder1,2,4,5,6
1Radiology, University of Wisconsin-Madison, Madison, WI, United States, 2Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 3Electrical and Computer Engineering, University of Wisconsin-Madison, Madison, WI, United States, 4Biomedical Engineering, University of Wisconsin-Madison, Madison, WI, United States, 5Medicine, University of Wisconsin-Madison, Madison, WI, United States, 6Emergency, University of Wisconsin-Madison, Madison, WI, United States Keywords: Data Processing, Artifacts, CSE-MRI In this work we develop a confidence map algorithm to identify reliable estimates of PDFF and R2* for CSE-MRI in the liver. Reliability of estimates is evaluated based on a normalized root-mean-square-error (NRMSE) between measured and model signals. Threshold values of the NRMSE are determined by using the Cramér–Rao Lower Bound. Monte-Carlo simulations and clinical CSE-MRI successfully validate the proposed method. |
| 09:19 | 0077.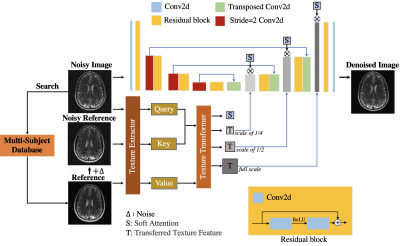 |
A Novel Cross-Subject Transformer Denoising Method
Shoujin Huang1, Sixing Liu1, Lifeng Mei1, Chenhui Tang1, Ed X Wu2,3, and Mengye Lyu1
1College of Health Science and Environmental Engineering, ShenZhen Technology University, Shenzhen, China, 2Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 3Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China Keywords: Data Processing, Modelling, Deep learning, Denoise In this work, we propose a new denoising method named Cross-Subject Transformer Denoising (CSTD), which transfers the texture of a reference image retrieved from a large database to the noisy image with soft attention mechanisms. The experiments on the fastMRI dataset with various noise levels show that our method is likely superior to many competing denoising algorithms including current the state-of-the-art NAFNet. Moreover, our method exhibits excellent generalizability when directly applied to in-vivo low-field data without retraining. Due to the flexibility, the method is expected to have a wide range of applications. |
| 09:27 | 0078.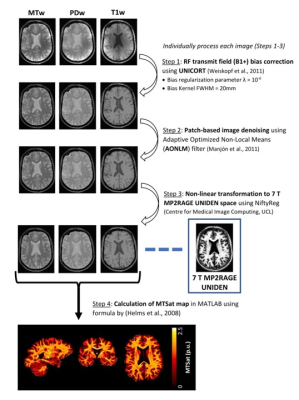 |
Optimized 7 T Cortical Magnetization Transfer Saturation Imaging: Application to Cortical Myelin Imaging in Multiple Sclerosis
Risavarshni Thevakumaran1,2, Marcus Couch2,3,4, Sridar Narayanan2,3, Douglas Arnold 2,3, and David Rudko1,2,3
1Department of Biological and Biomedical Engineering, McGill University, Montreal, QC, Canada, 2McConnell Brain Imaging Centre, Montreal Neurological Institute and Hospital, Montreal, QC, Canada, 3Department of Neurology and Neurosurgery, McGill University, Montreal, QC, Canada, 4Siemens Healthcare Limited, Montreal, QC, Canada Keywords: Data Processing, High-Field MRI, Quantitative MRI We designed an MRI protocol for rapid, high-resolution human cortical MTsat (csMTsat) imaging at 7 T. Our approach leveraged the PTx capability of the 7 T Siemens Terra MRI system. Further, we developed a post-processing scheme that reduces residual B1+ field bias and noise in csMTsat maps for imaging cortical myelin in the human cortex at high-resolution, which can be applied to study cortical demyelination in Multiple Sclerosis. |
09:35 |
0079.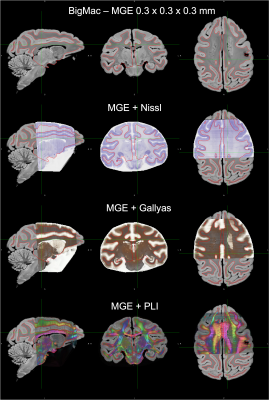 |
The joint registration of multiple microscopy contrasts to MRI in the BigMac dataset
Istvan N Huszar1, Silei Zhu2, Adele Smart2, Saad Jbabdi2, Karla L Miller2, and Amy F.D. Howard2
1Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 2University of Oxford, Oxford, United Kingdom Keywords: Data Processing, Data Processing, registration, microscopy, histology, postmortem, macaque We built a novel pipeline to register 290 high-resolution multi-contrast microscopy sections in the BigMac dataset to post-mortem MRI with sub-millimetre accuracy. The MRI-microscopy alignment is optimised over all microscopy sections jointly, to achieve high-quality co-registration whilst ensuring the slide order and separation is consistent with how the tissue was sectioned. We demonstrate the accuracy of the registration by overlaying FreeSurfer-estimated grey-white matter contours on orthogonal sections of 3D volumetric reconstructions of each microscopy modality. Our pipeline provides an integrated workflow for diffusion-microscopy studies using the BigMac dataset, which is openly available via the Oxford Digital Brain Bank. |
| 09:43 | 0080.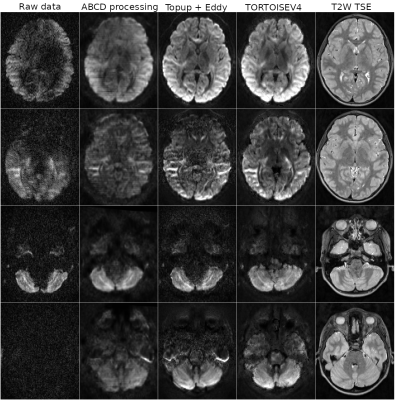 |
TORTOISE V4: ReImagining the NIH Diffusion MRI Processing Pipeline
M. Okan Irfanoglu1, Amritha Nayak1,2, Paul Taylor3, and Carlo Pierpaoli1
1QMI/NIBIB, National Institutes of Health, Bethesda, MD, United States, 2Henry Jackson Foundation, Bethesda, MD, United States, 3Scientific and Statistical Computing Core, NIMH, National Institutes of Health, Bethesda, MD, United States Keywords: Data Processing, Diffusion/other diffusion imaging techniques The processing needs for diffusion MRI data have evolved over the years with data sizes getting larger, diffusion sensitization going higher. Large multi-site studies, especially on "uncooperative subjects" such as young children or patients with movement disorders increased the necessity for dMRI processing pipelines that are fast, robustly capable of handling a variety of artifacts/distortions, and that have summary reporting capabilities that can pinpoint problematic data. The NIH Diffusion MRI processing pipeline, TORTOISE, has been reimagined, redesigned and and significantly enriched to satisfy these processing needs. |
| 09:51 | 0081.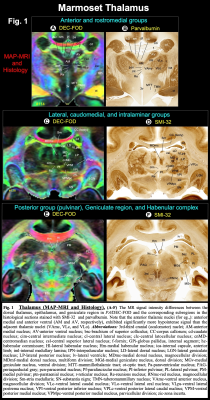 |
Multimodal anatomical mapping of subcortical regions in Marmoset monkeys using ultra-high resolution MAP-MRI and multiple histological stains
Kadharbatcha S Saleem1, Alexandru V Avram1, Daniel Glen2, Cecil Chern-Chyi Yen3, Vincent Schram4, and Peter J Basser1
1Section on Quantitative Imaging and Tissue Sciences (SQITS), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD-NIH), Bethesda, MD, United States, 2Scientific and Statistical Computing Core, National Institute of Mental Health (NIMH-NIH), Bethesda, MD, United States, 3Neuroimaging core, National Institute of Neurological Disorders and Stroke (NINDS-NIH), Bethesda, MD, United States, 4Microscopy and Imaging Core (MIC), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD-NIH), Bethesda, MD, United States Keywords: Visualization, Ex-Vivo Applications, MAP-MRI, subcortical, histology, Marmoset, and diffusion MRI Despite its importance as a model for human brain development and neurological disorders, the marmoset lacks a comprehensive MRI-histology-based parcellation of subcortical regions. Here, we mapped the subcortical areas of the marmoset brain in three dimensions (3D) using ultra-high resolution MAP-MRI, T2w, and MTR imaging at 7T, combined with histological stains of the same brain. Our results demonstrate that MAP-MRI can delineate cytoarchitectonic subregions of many deep brain structures observed with histology. Tracing and validating these important brain regions in 3D are imperative for neurosurgical planning, navigation of deep brain stimulation probes, and establishing brain structure-function relationships. |
09:59 |
0082.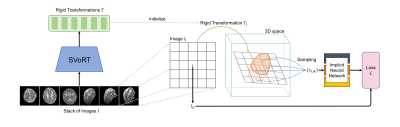 |
A Motion-Robust Slice-to-Volume Reconstruction Framework for Fetal Brain MRI
Junshen Xu1, Daniel Moyer2, Borjan Gagoski3, P. Ellen Grant3,4, Polina Golland5, Juan Eugenio Iglesias4,5,6,7, and Elfar Adalsteinsson1
1Massachusetts Institute of Technology, Cambridge, MA, United States, 2Computer Science, Vanderbilt University, Nashville, TN, United States, 3Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 4Harvard Medical School, Boston, MA, United States, 5Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, United States, 6Centre for Medical Image Computing, Department of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom, 7Athinoula A. Martinos Center for Biomedical Imaging, Cambridge, MA, United States Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence Volumetric reconstruction of fetal brains from multiple stacks of MR slices is challenging due to severe subject motion and image artifacts. We propose a deep learning method to solve the slice-to-volume reconstruction problem in two stages. First, a Transformer network is used to correct motion between slices by registering the input slices to a 3D canonical space. Second, an implicit neural network reconstructs the 3D volume by learning a continuous 3D representation of the fetal brain from the 2D observations. Results show that our method achieves high reconstruction quality and outperforms existing state-of-the-art methods in presence of severe fetal motion. |
| 10:07 | 0083.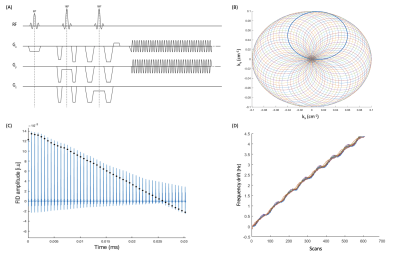 |
Characterising Magnetic Field Drift in Rosette-MRSI Data in-vivo
Sneha Vaishali Senthil1,2, Brenden Toshihide Kadota1, Peter Truong1, and Jamie Patrick Near1
1Medical Biophysics, Sunnybrook Research Institute, Toronto, ON, Canada, 2Integrated Program in Neuroscience, McGill University, Montreal, QC, Canada Keywords: Data Processing, Spectroscopy, Frequency drift correction, MRSI Field drift correction is an important post-processing step in MRS and MRSI that yields considerable improvements in spectral quality and metabolite quantification. While several drift correction methods have been proposed in single-voxel MRS, drift correction in MRSI is more challenging due to the application of phase encoding gradients. In such cases, separately acquired navigator echoes are needed. Here, we demonstrate the use of self-navigating Rosette-MRSI trajectories for characterising scanner magnetic field drift using time-domain spectral registration in-vivo. Results from the study showed substantial increases in SNR and spectral quality of the acquired data. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.