Oral Session
ML/AI Emerging Algorithms & Methodologies
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| 13:45 |
0154.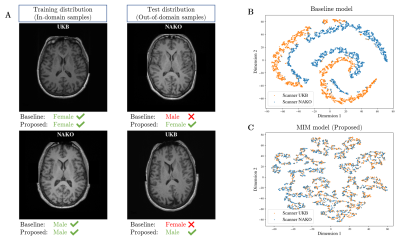 |
Avoiding shortcut-learning by mutual information minimization in
deep learning-based MR image processing
Louisa Fay1,2,
Bin Yang2,
Sergios Gatidis1,3,
and Thomas Kuestner1
1Medical Image and Data Analysis (MIDAS.lab), Department of Diagnostic and Interventional Radiology, University Hospital of Tuebingen, Tuebingen, Germany, 2Institute of Signal Processing and System Theory, University of Stuttgart, Stuttgart, Germany, 3Max Planck Institute for Intelligent Systems, Tuebingen, Germany Keywords: Machine Learning/Artificial Intelligence, Data Analysis Deep Learning methods can detect patterns in data such as MR images but are incapable of determining causal relationships. However, causal understanding is crucial in medical applications, since the presence of confounders (e.g. scan conditions) obscure the causal relationship and create spurious-correlations. State-of-the-art models purely rely on correlated patterns which can result in wrong conclusions or diagnoses when spurious-correlations change (e.g. new scanner). We propose a deep learning framework that is robust in the presence of spurious-correlations by decreasing mutual information between learned features of MR images and leads to improved performance under distribution shifts. |
| 13:53 |
0155. |
Adaptive convolution for including a-priori information in deep
learning models for quantitative susceptibility mapping
Simon Graf1,
Nora Küchler1,
Walter Wohlgemuth1,
and Andreas Deistung1
1University Hospital Halle (Saale), Halle (Saale), Germany Keywords: Machine Learning/Artificial Intelligence, Quantitative Susceptibility mapping Deep learning models used for solving dipole inversion of quantitative susceptibility mapping typically lack an integration of a-priori information. We show for the first time that information of voxel-size and field-of-view orientation with respect to B0 can be incorporated into network models by deploying adaptive convolution. Various network models were trained for 170 epochs to solve dipole inversion on synthetic data with arbitrary orientation and voxel-sizes. Adaptive convolution models outperform conventional models in computing susceptibility maps from arbitrarily oriented field distributions with anisotropic voxel sizes and allows a reduction of training time. |
14:01 |
0156.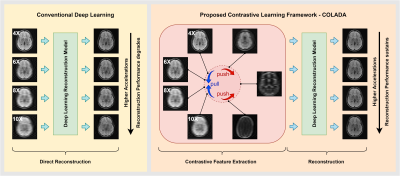 |
COLADA: Contrastive Learning for highly accelerated MR Image
Reconstruction
Mevan Ekanayake1,2,
Zhifeng Chen1,3,
Kamlesh Pawar1,
Gary Egan1,4,
Mehrtash Harandi2,
and Zhaolin Chen1,3
1Monash Biomedical Imaging, Monash University, Melbourne, Australia, 2Department of Electrical and Computer Systems Engineering, Monash University, Melbourne, Australia, 3Department of Data Science and AI, Monash University, Melbourne, Australia, 4School of Psychological Sciences, Monash University, Melbourne, Australia Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, self-supervised learning, contrastive learning, accelerated reconstruction, fastMRI Most deep learning methods for MR image reconstruction heavily depend on supervised learning on fully sampled reference data, hence, lack generalizability on out-of-distribution inputs such as higher accelerations. To reduce the models’ dependency on fully sampled reference data and to leverage large cohorts of undersampled MR measurements, we propose a self-supervised framework that extracts contrastive features between different accelerations of a given scan and the rest of the scans in the dataset, which we then utilize for the downstream reconstruction task. Our quantitative and qualitative analysis demonstrates the superiority of the proposed framework for highly accelerated MR image reconstruction. |
| 14:09 |
0157.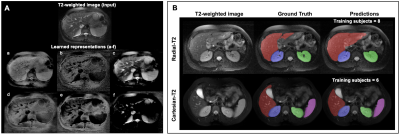 |
Reducing annotation burden in MR segmentation: A novel
contrastive learning loss with multi-contrast constraints on
local representations
Lavanya Umapathy1,2,
Taylor Brown2,3,
Mark Greenhill2,3,
J'rick Lu2,3,
Diego Martin4,
Maria Altbach2,
and Ali Bilgin1,2,5,6
1Department of Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States, 2Department of Medical Imaging, University of Arizona, Tucson, AZ, United States, 3College of Medicine, University of Arizona, Tucson, AZ, United States, 4Department of Radiology, Houston Methodist Hospital, Houston, TX, United States, 5Program in Applied Mathematics, University of Arizona, Tucson, AZ, United States, 6Department of Biomedical Engineering, University of Arizona, Tucson, AZ, United States Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Representational Learning The availability of limited labeled data motivates the use of self-supervised pretraining techniques for deep learning (DL) models. Here, we propose a novel contrastive loss that pushes/pulls local representations within an image based on representational constraints from co-registered multi-contrast MR images that share similar underlying parameters. For multi-organ segmentation tasks in T2-weighted images, pretraining a DL model using the proposed loss function with constraints from co-registered echo images from a radial TSE acquisition, can help reduce annotation burden by 60%. On two independent datasets, proposed pretraining improved Dice scores compared to random initialization and pretraining with conventional contrastive loss. |
| 14:17 |
0158.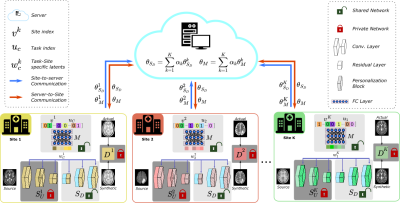 |
A Personalized Federated Learning Approach for Multi-Contrast
MRI Translation
Onat Dalmaz1,2,
Muhammad Usama Mirza1,2,
Gokberk Elmas1,2,
Muzaffer Ozbey1,2,
Salman UH Dar1,2,
Emir Ceyani3,
Salman Avestimehr3,
and Tolga Çukur1,2,4
1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center (UMRAM), Bilkent University, Ankara, Turkey, 3University of Southern California, Los Angeles, CA, United States, 4Neuroscience Program, Aysel Sabuncu Brain Research Center, Bilkent University, Ankara, Turkey Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Image Synthesis MRI contrast translation enables image imputation for missing sequences given acquired sequences in a multi-contrast protocol. Training of learning-based translation models requires access to large, diverse datasets that are challenging to aggregate centrally due to patient privacy risks. Federated learning (FL) is a promising solution that mitigates privacy concerns, but naive FL methods suffer from performance losses due to implicit and explicit data heterogeneities. Here, we introduce a novel FL-based personalized MRI translation method (pFLSynth) that effectively addresses implicit and explicit heterogeneity in multi-site datasets. FL experiments conducted on multi-contrast MRI datasets show the effectiveness of the proposed approach. |
14:25 |
0159.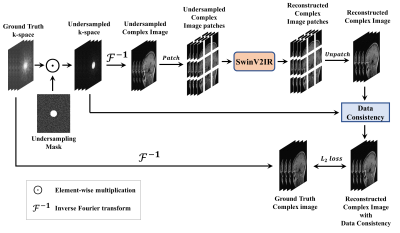 |
SwinV2-MRI: Accelerated Multi-Coil MRI Reconstruction using
Shifted Window Vision Transformers
Tahsin Rahman1,
Sergio D. Cabrera1,
and Ali Bilgin2
1Electrical and Computer Engineering, The University of Texas at El Paso, El Paso, TX, United States, 2Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction Vision transformers (ViT) are increasingly utilized in computer vision and have been shown to outperform CNNs in many tasks. In this work, we explore the use of Shifted Window (Swin) transformers for accelerated MRI reconstruction. Our proposed SwinV2-MRI architecture enables the use of multi-coil data and k-space consistency constraints with Swin transformers. Experimental results show that the proposed architecture outperforms CNNs even when trained on a limited dataset and without any pre-training. |
| 14:33 |
0160.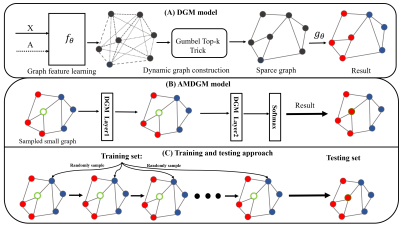 |
Developing a Graph Convolutional Network of Differentiable Graph
Module for Multi-Modal MRI Data: An Application to Parkinson's
Disease
Fanshi Li1,2,
Jun Li3,
Yifan Guo2,4,
Zhihui Wang1,2,
Zhilin Zhang2,4,
Xin Liu1,2,
Hairong Zheng1,2,
Yanjie Zhu1,2,
Liang Dong2,4,
and Haifeng Wang1,2 1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University of Chinese Academy of Sciences, Beijing, China, 3The Second People’s Hospital of Shenzhen, Shenzhen, China, 4Research Center for Medical AI, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China Keywords: Machine Learning/Artificial Intelligence, Parkinson's Disease With the ageing of the population, Parkinson's disease (PD) has presented a severe challenge to public health. Here, a deep-learning framework named the AMDGM model was proposed to predict PD patients at an early stage. Firstly, multi-modal image-based models were respectively generated using the AMDGM model. Then, a weighted ensemble network was created as the final model. The proposed method achieved the best AUC performance of 0.872 in the testing cohort, better than others. And the proposed method can predict PD patients early to help clinical radiologists formulate more targeted treatments in the future. |
| 14:41 |
0161.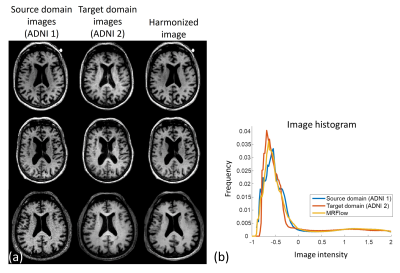 |
MRFlow: Flow-based neural network for MR image harmonization
Hwihun Jeong1,
Dong Un Kang1,
Jiye Kim1,
and Jongho Lee1
1Department of electrical and computer engineering, Seoul national university, Seoul, Korea, Republic of Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence We propose MRFlow, which is a normalizing flow-based neural network for the MRI harmonization framework. With the normalizing flow trained only with the target domain (e.g., 3T image) data, we harmonize the image from the source domain (e.g., 1.5T image) to the target domain by alternately reducing the norm of the latent variable and increasing similarity between the source domain and harmonized images. When MRFlow is applied to synthesized source domain images, the harmonized images showed lower errors than the source domain images. In the prospective study, the harmonized images became more similar to the target domain images after MRFlow. |
| 14:49 |
0162.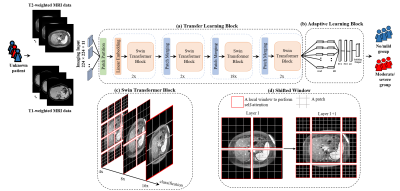 |
A transformer-based framework for liver stiffness classification
using multi-modality body MRI in children and adults
Redha Ali1,
Hailong Li1,
Huixian Zhang1,
Wen Pan2,
Scott B. Reeder3,
David T. Harris3,
William R. Masch4,
Anum Alsam4,
Krishna P. Shanbhogue5,
Anas Bernieh6,
Sarangarajan Ranganathan6,
Nehal A. Parikh6,
Jonathan R. Dillman1,
and Lili He1
1Department of Radiology, Cincinnati children's hospital medical center, Cincinnati, OH, United States, 2Department of Radiology, Cincinnati children's hospital medical center, 45429, OH, United States, 3University of Wisconsin-Madison, Madison, WI, United States, 4Michigan Medicine, University of Michigan, Ann Arbor, MI, United States, 5New York University Langone Health, New York, NY, United States, 6Cincinnati children's hospital medical center, Cincinnati, OH, United States Keywords: Machine Learning/Artificial Intelligence, Liver Magnetic resonance elastography (MRE) provides a noninvasive method to quantify liver stiffening, a surrogate biomarker for monitoring liver fibrosis. However, the availability of MRE remains limited, especially outside the United States, in part due to cost. This study aims to develop a deep learning-based approach for stratifying liver stiffness using multiparametric MRI images from pediatric and adult patients from multiple sites. We performed multi-site ten-fold cross-validation and achieved an AUROC of 0.80 for liver stiffness stratification. These results demonstrate that our proposed deep learning model may provide a means for categorical estimation of liver stiffening without dedicated elastography. |
| 14:57 |
0163.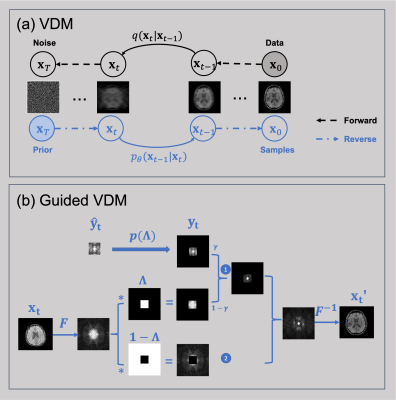 |
MR image super-resolution via a variational diffusion model
Wei Jiang1,
Yang Gao1,
and Hongfu Sun1
1University of Queensland, Brisbane, Australia Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, MRI, diffusion models High-resolution MR images are advantageous for medical diagnosis. However, they may require longer scan time and more powerful hardware. Significant effort has been made to carry out super-resolution methods to synthesize higher-resolution MR images from lower-resolution acquisitions using deep learning approaches. However, most current methods are biased. In this work, we propose a new super-resolution method based on the state-of-the-art generative model (i.e., variational diffusion model), using the lower-resolution K-space measurements as a condition for guiding the super-resolution process. This unsupervised approach achieved high performance for reconstructing MR images from arbitrary lower resolutions without retraining the model. |
| 15:05 |
0164.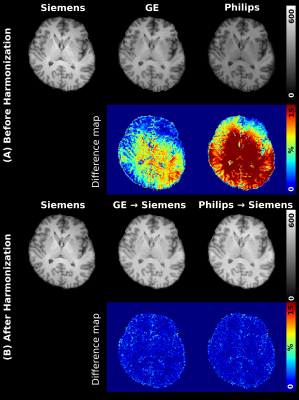 |
Bloch-informed neural network for multi-scanner MR Harmonization
Gawon Lee1,
Jun-Hyeok Lee1,
Dong Hye Ye2,
and Se-Hong Oh1
1Biomedical Engineering, Hankuk University of Foreign Studies, Yongin-si, Korea, Republic of, 2Computer Science, Georgia State University, Atlanta, GA, United States Keywords: Machine Learning/Artificial Intelligence, Brain, Data Harmonization For multi-site research in MR studies, a harmonization process is necessary. Due to the lack of paired dataset for harmonization, the unsupervised-based learning has been suggested. However, this method is susceptible to causing the error in anatomical information and performing well in the unseen domain. Thus, we suggest a deep neural network for multi-site harmonization based on MR signal physics with the paired traveling dataset. We implemented Quantitative Maps Generators and Denoising Network with the Bloch Equation module. We proved that using the Bloch Equation enhances the accuracy of harmonization. |
15:13 |
0165. |
Federated Learning for Utilizing Multi-Institutional Prostate
MRI with Diverse Histopathology
Abhejit Rajagopal1,
Katya Redekop2,
Anil Kemisetti1,
Rishi Kulkarni3,
Steven Raman3,
Karthik Sarma3,
Kirti Magudia4,
Corey Arnold2,3,
and Peder Larson1 1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Electrical Engineering, UCLA, Los Angeles, CA, United States, 3Radiology, UCLA, Los Angeles, CA, United States, 4Radiology, Duke University, Durham, NC, United States Keywords: Machine Learning/Artificial Intelligence, Cancer, federated learning Prostate cancer screening and diagnosis from MRI is extremely challenging, and current machine learning algorithms suffer in cross-institutional generalizability. Federated learning is a way to alleviate these issues by combining multi-center data without aggregating or homogenizing data. To enable this for prototype-stage algorithms, we introduce FLtools, a lightweight python library with re-usable federated learning components available freely at https://federated.ucsf.edu. We use this federated learning system to train a 3D UCNet on bi-parametric MRI and paired prostate biopsy data from two University of California hospitals, demonstrating dramatic improvements in cross-site generalization accuracy in clinically-significant lesion classification. |
| 15:21 |
0166.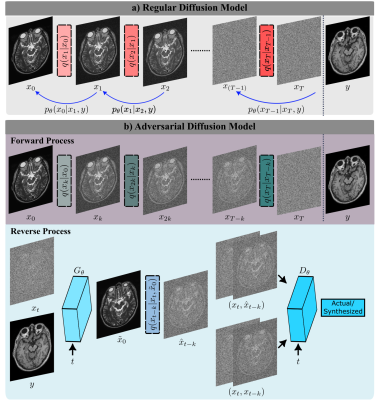 |
Adversarial Diffusion Probabilistic Models for Unpaired MRI
Contrast Translation
Muzaffer Ozbey1,2,
Onat Dalmaz1,2,
Salman UH Dar1,2,
Hasan Atakan Bedel1,2,
Şaban Öztürk1,2,
Alper Güngör1,2,
and Tolga Çukur1,2,3
1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center (UMRAM), Bilkent University, Ankara, Turkey, 3Neuroscience Program, Aysel Sabuncu Brain Research Center, Bilkent University, Ankara, Turkey Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence Synthesis of missing contrasts in an MRI protocol via translation from acquired contrasts can reduce costs associated with prolonged exams. Current learning-based translation methods are predominantly based on generative adversarial networks (GAN) that implicitly characterize the distribution of the target contrast, with limits fidelity of synthesized images. Here we present SynDiff, a novel conditional adversarial diffusion model for computationally efficient, high-fidelity contrast translation. SynDiff enables training on unpaired datasets, thanks to its cycle-consistent architecture with coupled diffusion processes. Demonstrations on multi-contrast MRI datasets indicate the superiority of SynDiff against competing GAN and diffusion models. |
| 15:29 |
0167.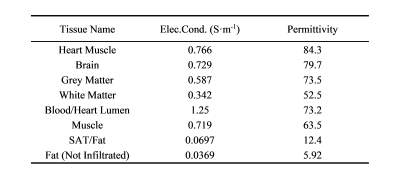 |
A Machine Learning-Based Prediction of RF Transfer Function of
Active Implantable Medical Devices in Homogeneous and
Heterogeneous Tissue
Mingjuan Ma1,
Hexuan Shi1,
and Aiping Yao1 1Lanzhou University, Lanzhou, China Keywords: Bioeffects & Magnetic Fields, Safety Both the Tier-3 and Tier-4 methods proposed in ISO/TS 10974 for assessing the radiofrequency safety of active implantable medical devices have limitations in practice. The transfer function proposed in the Tier-3 method can only be measured individually for each implant and only for homogeneous tissue environments, while Tier-4 requires significant high-speed computing resources. In this study, machine learning algorithms are proposed to predict the transfer function both in homogeneous and heterogeneous tissue environment. The results show that this approach is feasible and performs well in the prediction task. |
| 15:37 |
0168.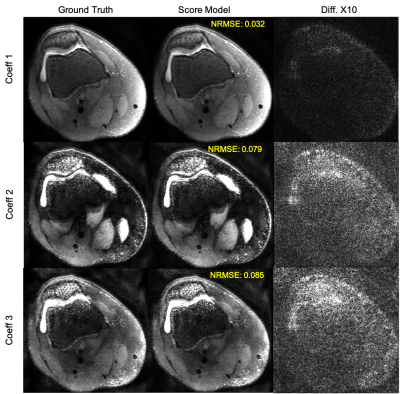 |
Multi-Contrast 3D Fast Spin-Echo T2 Shuffling Reconstruction
with Score-Based Deep Generative Priors
Sidharth Kumar1,
Asad Aali1,
and Jonathan I Tamir1,2,3
1Chandra Family Department of Electrical and Computer Engineering, University of Texas at Austin, Austin, TX, United States, 2Oden Institute for Computational Engineering and Sciences, University of Texas at Austin, Austin, TX, United States, 3Department of Diagnostic Medicine, University of Texas at Austin, Austin, TX, United States Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction Score-based generative modeling has emerged as a powerful tool for modeling image priors and has recently been used to solve ill-posed inverse problems in various domains including MRI reconstruction. Here we extend the framework to reconstruct multi-contrast 3D fast spin-echo (FSE), i.e. T2 Shuffling data. This is achieved by constraining the posterior sampling reconstruction to a low-dimensional subspace and training a score model on images from this subspace. We demonstrate a proof-of-principal reconstruction of data with no model mismatch, i.e. generated from the forward model. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.