Oral Session
ML/AI for Analysis, Post-Processing, Disease Diagnosis & Prediction
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| 16:00 |
1368.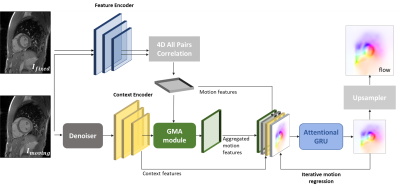 |
Attention-guided network for image registration of accelerated
cardiac CINE
Aya Ghoul1,
Kerstin Hammernik2,3,
Patrick Krumm4,
Sergios Gatidis1,5,
and Thomas Küstner1
1Medical Image And Data Analysis (MIDAS.lab), Department of Diagnostic and Interventional Radiology, University Hospital of Tuebingen, Tuebingen, Germany, 2Lab for AI in Medicine, Technical University of Munich, Munich, Germany, 3Department of Computing, Imperial College London, London, United Kingdom, 4Department of Radiology, University Hospital of Tuebingen, Tuebingen, Germany, 5Max Planck Institute for Intelligent Systems, Tuebingen, Germany Keywords: Machine Learning/Artificial Intelligence, Motion Correction, Image registration, Image Reconstruction Motion-resolved reconstruction methods permit for considerable acceleration for cardiac CINE acquisition. Solving for the non-rigid cardiac motion is computationally demanding, and even more challenging in highly accelerated acquisitions, due to the undersampling artifacts in image domain. Here, we introduce a novel deep learning-based image registration network, GMA-RAFT, for estimating cardiac motion from accelerated imaging. A transformer-based module enhances the iterative recurrent refinement of the estimated motion by introducing structural self-similarities into the decoded features. Experiments on Cartesian and radial trajectories demonstrate superior results compared to other deep learning and state-of-the-art baselines in terms of motion estimation and motion-compensated reconstruction. |
| 16:08 |
1369.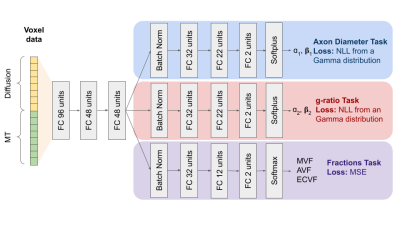 |
A transfer learning approach to predict Axon Diameter and
g-ratio distributions from MRI Data
Gustavo Chau Loo Kung*1,2,
Emmanuelle M. M. Weber*2,
Juliet Knowles3,
Ankita Batra3,
Lijun Ni3,
and Jennifer McNab2
1Bioengineering Department, Stanford University, Stanford, CA, United States, 2Radiology Department, Stanford University, Stanford, CA, United States, 3Neurology Department, Stanford University, Stanford, CA, United States Keywords: Machine Learning/Artificial Intelligence, Microstructure, Histology, Diffusion Imaging, g-ratio, axon diameter To better establish the influence of histological features on the MRI signal, we present a multi-task neural network trained to predict parametrized microstructural distributions (axon diameters and g-ratios) from diffusion and magnetization transfer MRI data. To begin, we trained the model using histologically-derived synthetic MRI data before applying transfer learning by fine tuning on empirical data. Our initial results on both synthetic and empirical ex vivo mouse brain MRI data demonstrate the feasibility of this approach. |
| 16:16 |
1370.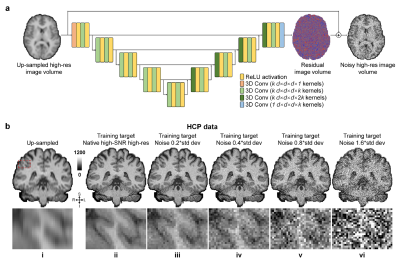 |
SRNR: Training neural networks for Super-Resolution MRI using
Noisy high-resolution Reference data
Jiaxin Xiao1,
Zihan Li2,
Berkin Bilgic3,4,
Jonathan R. Polimeni3,4,
Susie Huang3,4,
and Qiyuan Tian3,4
1Department of Electronic Engineering, Tsinghua University, Beijing, China, 2Department of Biomedical Engineering, Tsinghua University, Beijing, China, 3Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Charlestown, MA, United States, 4Harvard Medical School, Boston, MA, United States Keywords: Machine Learning/Artificial Intelligence, Data Analysis Neural network (NN) based approaches for super-resolution MRI typically require high-SNR high-resolution reference data acquired in many subjects, which is time consuming and a barrier to feasible and accessible implementation. We propose to train NNs for Super-Resolution using Noisy Reference data (SRNR), leveraging the mechanism of the classic NN-based denoising method Noise2Noise. We systematically demonstrate that results from NNs trained using noisy and high-SNR references are similar for both simulated and empirical data. SRNR suggests a smaller number of repetitions of high-resolution reference data can be used to simplify the training data preparation for super-resolution MRI. |
| 16:24 |
1371.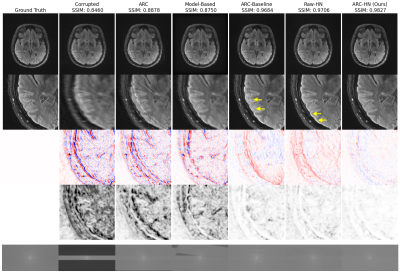 |
Motion-Aware Neural Networks Improve Rigid Motion Correction of
Accelerated Segmented Multislice MRI
Nalini M. Singh1,2,
Malte Hoffmann3,4,
Elfar Adalsteinsson2,5,6,
Bruce Fischl2,3,4,
Polina Golland*1,5,6,
Adrian V. Dalca*1,3,4,
and Robert Frost*3,4
1Computer Science and Artificial Intelligence Laboratory (CSAIL), MIT, Cambridge, MA, United States, 2Harvard-MIT Health Sciences and Technology, MIT, Cambridge, MA, United States, 3Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 4Department of Radiology, Harvard Medical School, Boston, MA, United States, 5Department of Electrical Engineering and Computer Science, MIT, Cambridge, MA, United States, 6Institute for Medical Engineering and Science, MIT, Cambridge, MA, United States Keywords: Machine Learning/Artificial Intelligence, Motion Correction, Image Reconstruction, Deep Learning We demonstrate a deep learning approach for fast retrospective intraslice rigid motion correction in segmented multislice MRI. A hypernetwork uses auxiliary rigid motion parameter estimates to produce a reconstruction network based on the motion parameters that are specific to the input image. This strategy produces higher quality reconstructions than those produced by model-based techniques or by networks that do not use motion estimates. Further, this approach mitigates sensitivity to misestimation of the motion parameters. |
| 16:32 |
1372.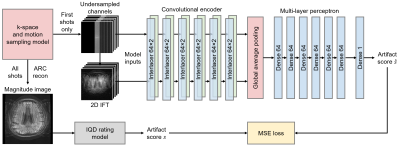 |
Can we predict motion artifacts in clinical MRI before the scan
completes?
Malte Hoffmann1,2,
Nalini M Singh3,4,
Adrian V Dalca1,2,3,
Bruce Fischl1,2,3,4,
and Robert Frost1,2
1Department of Radiology, Harvard Medical School, Boston, MA, United States, 2Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 3Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, United States, 4Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States Keywords: Machine Learning/Artificial Intelligence, Artifacts, deep learning, AI-guided radiology, neuroimaging, computer vision Subject motion remains the major source of artifacts in magnetic resonance imaging (MRI). Motion correction approaches have been successfully applied in research, but clinical MRI typically involves repeating corrupted acquisitions. To alleviate this inefficiency, we propose a deep-learning strategy for training networks that predict a quality rating from the first few shots of accelerated multi-shot multi-slice acquisitions, scans frequently used for neuroradiological screening. We demonstrate accurate prediction of the scan outcome from partial acquisitions, assuming no further motion. This technology has the potential to inform the operator's decision on aborting corrupted scans early instead of waiting until the acquisition completes. |
| 16:40 |
1373.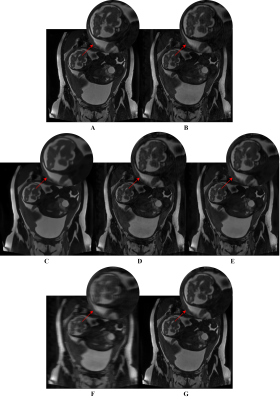 |
A Deep Learning Method to Remove Motion Artifacts in Fetal MRI
Adam Lim1,2,
Justin Lo1,2,
Matthias Wagner3,
Birgit Ertl-Wagner3,4,
and Dafna Sussman1,2,5
1Department of Electrical, Computer and Biomedical Engineering, Toronto Metropolitan University, Toronto, ON, Canada, 2Institute for Biomedical Engineering, Science and Technology (iBEST), Toronto Metropolitan University and St. Michael’s Hospital, Toronto, ON, Canada, 3Division of Neuroradiology, The Hospital for Sick Children, Toronto, ON, Canada, 4Department of Obstetrics and Gynecology, University of Toronto, Toronto, ON, Canada, 5Department of Medical Imaging, University of Toronto, Toronto, ON, Canada Keywords: Machine Learning/Artificial Intelligence, Artifacts, Deep Learning, Generative Adversarial Network, Image Denoising Motion artifacts are a common issue in fetal MR imaging that limit the visibility of essential fetal anatomy. In such cases, the sequence acquisition must be repeated in order for an accurate diagnosis. This study introduces a deep learning approach utilizing a Generative Adversarial Network (GAN) framework for removing motion artifacts in fetal MRIs. Results exceeded current state-of-the-art methods by achieving an average SSIM of 93.7%, and PSNR of 33.5dB. The presented network demonstrates rapid and accurate results that can be advantageous in clinical use. |
16:48 |
1374.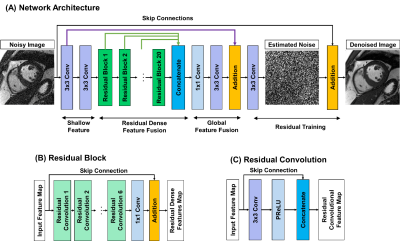 |
Cardiac MR Denoising Inline Neural Network (CaDIN).
Siyeop Yoon1,
Salah Assana1,
Manuel A. Morales1,
Julia Cirillo1,
Patrick Pierce1,
Beth Goddu1,
Jennifer Rodriguez1,
and Reza Nezafat1 1Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA, United States Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction The diagnostic confidence in the interpretation of cardiac MR scans can be improved by enhancing the signal-to-noise ratio (SNR). Traditional image denoising has been studied extensively to improve SNR in cardiac MRI, but with limited success due to the resulting blurring. In this study, we sought to develop and evaluate cardiac MR denoising inline neural network (CaDIN) for improving SNR in cardiac MRI. |
16:56 |
1375.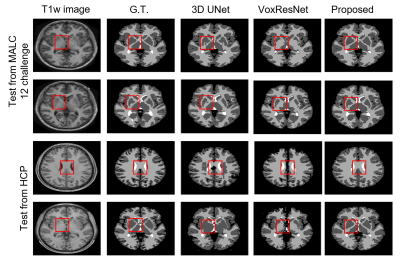 |
Improved Bayesian Brain MR Image Segmentation by Incorporating
Subspace-Based Spatial Prior into Deep Neural Networks
Yunpeng Zhang1,
Huixiang Zhuang1,
Ziyu Meng1,
Ruihao Liu1,2,
Wen Jin2,3,
Wenli Li1,
Zhi-Pei Liang2,3,
and Yao Li1
1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States Keywords: Machine Learning/Artificial Intelligence, Segmentation Accurate segmentation of brain tissues is important for brain imaging applications. Learning the high-dimensional spatial-intensity distributions of brain tissues is challenging for classical Bayesian classification and deep learning-based methods. This paper presents a new method that synergistically integrate a tissue spatial prior in the form of a mixture-of-eigenmodes with deep learning-based classification. Leveraging the spatial prior, a Bayesian classifier and a cluster of patch-based position-dependent neural networks were built to capture global and local spatial-intensity distributions, respectively. By combining the spatial prior, Bayesian classifier, and the proposed networks, our method significantly improved the segmentation performance compared with the state-of-the-art methods. |
| 17:04 |
1376.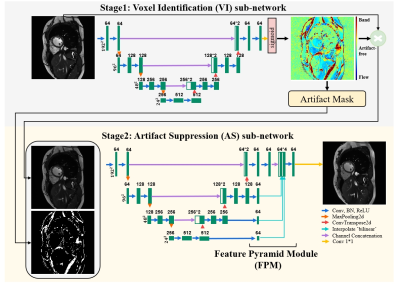 |
A dual-stage partially interpretable neural network for joint
suppression of bSSFP banding and flow artifacts in
non-phase-cycled cine imaging
Zhuo Chen1,
Juan Gao1,
Xin Tang1,
and Chenxi Hu1
1The Institute of Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University (SJTU), Shanghai, China Keywords: Machine Learning/Artificial Intelligence, Artifacts, Banding artifacts, Flow artifacts bSSFP cine imaging suffers from banding and flow artifacts in the region of off-resonance. Suppressing one kind of artifacts may evoke the other kind. For example, phase cycling suppresses banding artifacts, yet its acquisition at multiple frequency offsets often evokes flow artifacts. Here, we develop a partially interpretable neural network for jointly suppressing banding and flow artifacts without phase cycling. Based on a single cine image, the method generates an artifact-corrected image and a voxel-identity map, which guides the artifact suppression and improves its interpretability. Preliminary investigation shows that the method reduces banding and flow artifacts without introducing new artifacts. |
| 17:12 |
1377.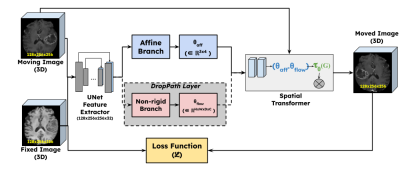 |
Non-rigid guided affine image registration in multi-contrast
brain MRI using deep networks with stochastic depth
Srivathsa Pasumarthi Venkata1 and
Ryan Chamberlain1
1Research & Development, Subtle Medical Inc, Menlo Park, CA, United States Keywords: Machine Learning/Artificial Intelligence, Brain, Image Registration Deep learning based affine registration is a fast and computationally efficient alternative to conventional iterative methods. However, existing solutions are not sensitive to local misalignments. We propose a non-rigid guided affine registration network with stochastic depth which was designed with an affine branch and an optional non-rigid branch. The probability of dropping the non-rigid branch was gradually increased over training epochs. During inference, the non-rigid branch was fully removed, thus making it a pure affine network guided by non-rigid transformations. Model training and quantitative evaluation was performed using a pre-registered multi-contrast brain MRI public dataset. |
17:20 |
1378.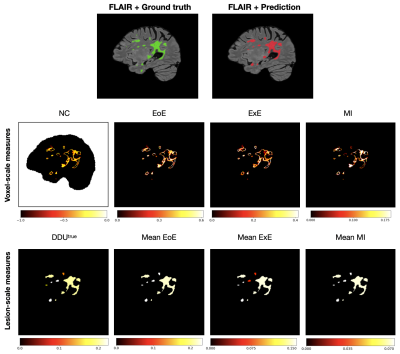 |
Towards Informative Uncertainty Measures for MRI Segmentation in
Clinical Practice: Application to Multiple Sclerosis
Nataliia Molchanova1,2,3,
Vatsal Raina3,4,
Francesco La Rosa5,
Andrey Malinin6,
Henning Müller3,
Mark Gales4,
Cristina Granziera7,
Mara Graziani3,8,
and Merirxell Bach Cuadra1,9
1Radiology department, Lausanne University Hospital (CHUV), Lausanne, Switzerland, 2Doctoral School of the Faculty of Biology and Medicine, University of Lausanne (UNIL), Lausanne, Switzerland, 3University of Applied Sciences of Western Switzerland, Sierre, Switzerland, 4University of Cambridge, Cambridge, United Kingdom, 5Icahn School of Medicine at Mount Sinai, New York, NY, United States, 6Shifts Project, Helsinki, Finland, 7University Hospital Basel, Basel, Switzerland, 8IBM Research Europe, Zurich, Switzerland, 9Center for Biomedical Imaging (CIBM), University of Lausanne, Lausanne, Switzerland Keywords: Machine Learning/Artificial Intelligence, Multiple Sclerosis, Machine learning/Artificial intelligence, Brain, Uncertainty estimation, Reliable AI We approach the problem of quantifying the degree of reliability of supervised deep learning models used by clinicians for automatic multiple sclerosis lesion segmentation on MRI. In particular, we quantify the correspondence of various uncertainty measures to the errors that a deep learning model makes in overall segmentation or lesion detection. The evaluation is done both on in- and out-of- domain datasets (40 and 99 patients respectively), and provides insights about the measures that can point clinicians to potential errors of an automatic algorithm regardless of the distributional shift. |
| 17:28 |
1379.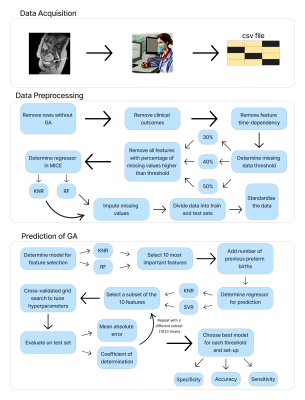 |
Predicting Gestational Age at Birth in the Context of Preterm
Birth Using Comprehensive Fetal MRI Acquisitions
Diego Fajardo-Rojas1,
Riine Heinsalu1,
Megan Hall2,
Mary Rutherford3,
Joseph Hajnal4,
Emma Robinson4,
Lisa Story2,
and Jana Hutter3
1School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom, 2Department of Women & Children's Health, Guy's and St Thomas' NHS Foundation Trust, London, United Kingdom, 3Department of Perinatal Imaging & Health, King's College London, London, United Kingdom, 4Department of Biomedical Engineering, King's College London, London, United Kingdom Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence The accurate prediction of preterm birth is a clinically crucial but challenging problem due to its complex aetiology. In this work, data from fetal anatomical and functional multi-organ MRI acquisitions are used to train Random Forests and Support Vector Machines to predict gestational age at delivery. These predictions are classified as 'term' or 'preterm'. The model with highest sensitivity, a Random Forest, achieved 0.85 sensitivity, 0.81 accuracy, 0.8 specificity, 1.99 weeks Mean Absolute Error, and 0.58 R2 score. This work proves the potential of Machine Learning models trained on anatomical and functional MRI data to predict gestational age at delivery. |
| 17:36 |
1380.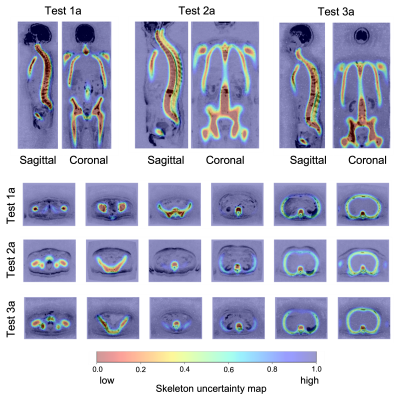 |
Uncertainty maps for training a deep learning model that
automatically delineates the skeleton from Whole-Body Diffusion
Weighted Imaging
Antonio Candito1,
Martina Torcè2,
Richard Holbrey3,
Alina Dragan1,
Christina Messiou1,
Nina Tunariu1,
Dow-Mu Koh1,
and Matthew D Blackledge1
1The Institute of Cancer Research, London, United Kingdom, 2Imperial College London, London, United Kingdom, 3Mint Medical, Heidelberg, Germany Keywords: Machine Learning/Artificial Intelligence, Segmentation Whole-Body Diffusion Weighted Imaging (WBDWI) requires automated tools that delineate malignant bone disease based on high b-value signal intensity, leading to state-of-the-art imaging biomarkers of response. As an initial step, we have developed an automated deep-learning pipeline that automatically delineates the skeleton from WBDWI. Our approach is trained on paired examples, where ground truth is defined through a set of weak labels (non-binary segmentations) derived from a computationally expensive atlas-based segmentation approach. The model showed on average a dice score, precision and recall between the manual and derived skeleton segmentations on test datasets of 0.74, 0.78, and 0.7, respectively. |
| 17:44 |
1381.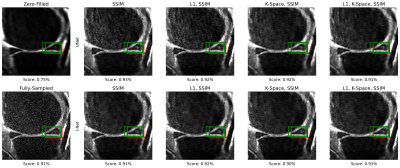 |
Towards Integrating DL Reconstruction and Diagnosis: Meniscal
Anomaly Detection Shows Similar Performance on Reconstructed and
Baseline MRI
Natalia Konovalova1,
Aniket Tolpadi1,2,
Felix Liu1,
Rupsa Bhattacharjee1,
Felix Gassert1,
Paula Giesler1,
Johanna Luitjens1,
Sharmila Majumdar1,
and Valentina Pedoia1
1Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States, 2Department of Bioengineering, University of California, Berkeley, Berkeley, CA, United States Keywords: Joints, Machine Learning/Artificial Intelligence Meniscal lesions are a common knee pathology, but pathology detection from MRI is usually evaluated on full-length acquisitions. We trained UNet and KIKI I-Net reconstruction algorithms with several loss function configurations, showing k-space losses are not required to obtain robust reconstructions. We trained and evaluated Faster R-CNN to detect meniscal anomalies, showing similar performance on R=8 reconstructions and fully-sampled images, demonstrating its utility as an assessment tool for reconstruction performance and indicating reconstructed images are viable for downstream clinical postprocessing tasks. |
| 17:52 |
1382.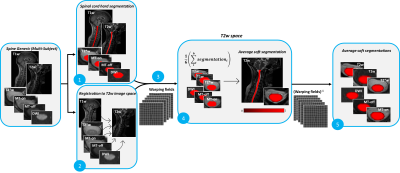 |
Contrast-agnostic segmentation of the spinal cord using deep
learning
Sandrine Bédard1,
Adrian El Baz1,
Uzay Macar1,
and Julien Cohen-Adad1,2,3,4
1NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montréal, QC, Canada, 2Functional Neuroimaging Unit, CRIUGM, University of Montreal, Montréal, QC, Canada, 3Mila - Quebec AI Institute, Montréal, QC, Canada, 4Centre de recherche du CHU Sainte-Justine, Université de Montréal, Montréal, QC, Canada Keywords: Machine Learning/Artificial Intelligence, Segmentation Several methods to segment the spinal cord have emerged over the past decade. However, they are dependent on the image contrast, resulting in differences of spinal cord cross-sectional area (CSA), a relevant biomarker in neurodegenerative diseases. We propose a novel method using deep learning that produces the same segmentation regardless of the MRI contrast. Moreover, the segmentation is “soft” (non-binary) and can therefore encode partial volume information. CSA computed with this contrast-agnostic soft segmentation method has lower intra- and inter-subject variability, making it particularly relevant for multi-center studies. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.