Oral Session
ML/AI New Ideas
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| 15:45 |
0979.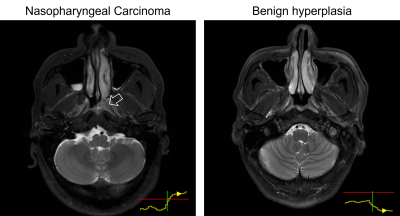 |
Explainable artificial intelligence for automatic detection of
early nasopharyngeal carcinoma on MRI
Lun M Wong1,
Qi-Yong H Ai1,2,
Tiffany Y So1,
Jacky WK Lam3,4,
and Ann D King1
1Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, Hong Kong, 2Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, Hong Kong, 3Department of Chemical Pathology, The Chinese University of Hong Kong, Hong Kong, Hong Kong, 4State Key Laboratory of Translational Oncology, The Chinese University of Hong Kong, Hong Kong, Hong Kong Keywords: Machine Learning/Artificial Intelligence, Screening, XAI, explainability, deep learning In this study, we propose a simple method to improve the explainability of artificial intelligence, specifically convolutional neural networks (CNNs), for the automatic detection of early nasopharyngeal carcinoma (NPC) on magnetic resonance imaging (MRI). We show a long-short-term-memory (LSTM) unit can be introduced into a CNN to read 3-dimensional medical image series. A risk curve can be extracted from the LSTM to visualize the “thought process” of the network when it reads through the input MRI slice-by-slice. This modification improves the explainability of the network without reducing performance for the early NPC detections of the original CNN. |
| 15:53 |
0980.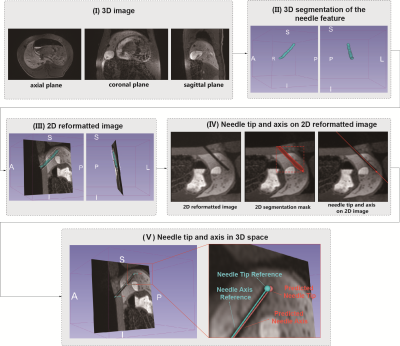 |
Deep Learning-Based Automatic Pipeline for 3D Needle
Localization on Intra-Procedural 3D MRI
Wenqi Zhou1,2,
Xinzhou Li1,2,
Fatemeh Zabihollahy1,
David S. Lu1,
and Holden H. Wu1,2
1Department of Radiological Sciences, UCLA, Los Angeles, CA, United States, 2Department of Bioengineering, UCLA, Los Angeles, CA, United States Keywords: Machine Learning/Artificial Intelligence, MR-Guided Interventions Needle localization in 3D images during MRI-guided interventions is challenging due to the complex structure of biological tissue and the variability in the appearance of needle features in in-vivo MR images. Deep learning networks such as the Mask Regional Convolutional Neural Network (R-CNN) could address this challenge by providing accurate needle feature segmentation in intra-procedural MR images. This work developed an automatic coarse-to-fine pipeline that combines 2.5D and 2D Mask R-CNN to leverage inter-slice information and localize the needle tip and axis in in-vivo intra-procedural 3D MR images. |
| 16:01 |
0981.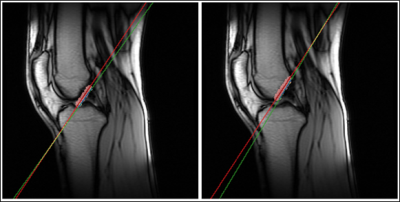 |
Intelligent Slice Placement for Oblique Coronal ACL plane
prescription for Knee MRI using Deep Learning
Apoorva Agarwal1,
Chitresh Bhushan2,
Desmond T.B. Yeo2,
Thomas Foo2,
Dawei Gui3,
Deepa Anand1,
Maggie Fung4,
Trevor Kolupar3,
Tisha Abraham1,
Sanjay NT1,
Kameswari Padmanabhan1,
Uday Patil1,
and Dattesh Shanbhag1
1GE Healthcare, Bengaluru, India, 2GE Global Research, Niskayuna, NY, United States, 3GE Healthcare, Waukesha, WI, United States, 4GE Healthcare, New York City, NY, United States Keywords: Machine Learning/Artificial Intelligence, MSK In clinical practice, oblique coronal and sagittal scans have been shown to be superior for visualization of ACL and assessment of ACL tears. In this work, we demonstrate a Deep Learning based method to obtain these complex scan planes (Coronal ACL and Coronal Blumensaat) directly from standard 2D tri-planar Localizers. Results on 487 test volumes show <0.6mm mean error for both planes, indicating suitability for clinical usage. |
| 16:09 |
0982.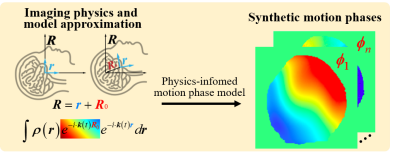 |
Physics-informed Deep Diffusion MRI Reconstruction: Break the
Data Bottleneck in Artificial Intelligence
Chen Qian1,
Yiting Sun1,
Zi Wang1,
Xinlin Zhang1,
Qinrui Cai1,
Taishan Kang2,
Boyu Jiang3,
Ran Tao3,
Zhigang Wu4,
Di Guo5,
and Xiaobo Qu1
1Department of Electronic Science, Biomedical Intelligent Cloud R&D Center, Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2Department of Radiology, Zhongshan Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, China, 3United Imaging Healthcare, Shanghai, China, 4Philips, Beijing, China, 5School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China Keywords: Machine Learning/Artificial Intelligence, Brain, Diffusion MR, Physics-informed Deep learning is widely employed in biomedical magnetic resonance image (MRI) reconstructions. However, accurate training data are unavailable in multi-shot interleaved echo planer imaging (Ms-iEPI) diffusion MRI (DWI) due to inter-shot motion. In this work, we propose a Physics-Informed Deep DWI reconstruction method (PIDD). For Ms-iEPI DWI data synthesis, a simplified physical motion model for motion-induced phase synthesis is proposed. Then, lots of synthetic phases are combined with a few real data to generate efficient training data. Extensive results show that, PIDD trained on synthetic data enables sub-second, ultra-fast, high-quality, and robust reconstruction with different b-values and undersampling patterns. |
| 16:17 |
0983.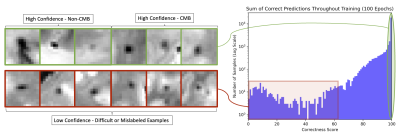 |
Ex-vivo microbleed detection in community-based older adults
using confidence aware learning
Grant Nikseresht1,
Arnold M. Evia2,
David A. Bennett2,
Julie A. Schneider2,
Gady Agam1,
and Konstantinos Arfanakis2,3
1Computer Science, Illinois Institute of Technology, Chicago, IL, United States, 2Rush Alzheimer's Disease Center, Rush University Medical Center, Chicago, IL, United States, 3Biomedical Engineering, Illinois Institute of Technology, Chicago, IL, United States Keywords: Machine Learning/Artificial Intelligence, Aging, Ex-Viov Applications, Neuro (Microbleeds) Detection of cerebral microbleeds (CMBs) on ex-vivo gradient echo MR images is an important task in MRI-pathology studies in older adults. The goal of this study was to develop a confidence-aware ex-vivo CMB detection algorithm that outputs interpretable probabilities and ranking of CMB candidates on brain MRI scans of community-based older adults. The present study demonstrates that training an ex-vivo CMB detection model with confidence-aware deep learning, a technique for improving confidence estimation and ordinal ranking of examples in classification models, improves detection performance and prediction interpretability. |
| 16:25 |
0984.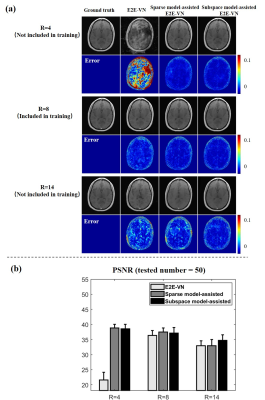 |
Model-Assisted Deep Learning-Based Reconstruction: Does the
Model Help?
Yue Guan1,
Yudu Li2,3,
Ruihao Liu1,2,
Ziyu Meng1,
Yao Li1,
Yiping P. Du1,
and Zhi-Pei Liang2,4
1School of Biomedical Engineering, Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3National Center for Supercomputing Applications, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction Deep learning-based image reconstruction has two known practical issues: (a) sensitivity to data perturbations, and (b) poor generalization. An approach to addressing these issues is to use classical signal models to assist/constrain deep learning. This paper performs a systematic analysis of the role of signal models in model-assisted deep learning-based reconstruction. Our results show that signal models (e.g., subspace model or sparse model) can substantially reduce the sensitivity of deep learning-based reconstruction to data perturbations; they can also help improve generalization capability. |
| 16:33 |
0985.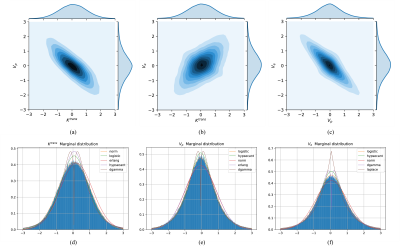 |
Pharmacokinetic Parameters’ Distribution Estimation with
Normalizing Flow in Dynamic Contrast-Enhanced Magnetic Resonance
Imaging
Ke Fang1,
Zejun Wang2,
Zhaowei Cheng1,
Bao Wang3,
Xinyu Jin1,
Yingchao Liu4,
and Ruiliang Bai5
1College of Information Science and Electronic Engineering, Zhejiang University, Hangzhou, China, 2Key Laboratory of Biomedical Engineering of Ministry of Education, College of Biomedical Engineering and Instrument Science, Zhejiang University, Hangzhou, China, 3Department of Radiology, Qilu Hospital of Shandong University, Jinan, China, 4Department of Neurosurgery, Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China, 5School of Medicine, Zhejiang University, Hangzhou, China Keywords: Machine Learning/Artificial Intelligence, Data Analysis The pharmacokinetic (PK) parameters extracted from the DCE-MRI provide valuable information but suffer from many sources of variability. Thus, the efficient and fast estimation of the distributions of these ambiguous PK parameters caused by variabilities could significantly improve the robustness and repeatability of DCE-MRI. The estimation of the PK parameters’ distributions provides a way to quantify the PK parameters’ values and variabilities simultaneously. In this study, we demonstrated the feasibility of the normalizing flow-based distribution estimation network (FPDEN) for PK parameters’ distribution estimation in DCE-MRI. |
| 16:41 |
0986.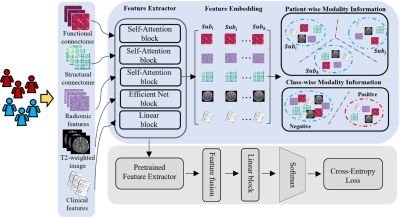 |
A novel deep multimodal contrastive network for early prediction
of cognitive deficits using multimodal MRI and clinical data
Zhiyuan Li1,2,
Hailong Li1,
Nehal Parikh3,
Jonathan Dillman1,
Anca Ralescu2,
and Lili He1
1Imaging Research Center, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Department of Computer Science, University of Cincinnati, Cincinnati, OH, United States, 3The Perinatal Institute, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States Keywords: Machine Learning/Artificial Intelligence, Brain Connectivity, Contrastive Learning, Cognitive Deficits Deep learning has shown promising results in early predicting cognitive deficits in very preterm infants using multimodal brain MRI data, acquired soon after birth. Prior methods focus on the feature fusion of different modalities but ignore the latent high-order feature similarity information. In this study, we propose a novel deep multimodal contrastive network using T2-weighted structural MRI (sMRI), diffusion tensor imaging (DTI), resting-state functional MRI (rsfMRI), and clinical data to predict later cognitive deficits. Our proposed model significantly improved predictive power compared to other peer models for early diagnosis of cognitive deficits. |
| 16:49 |
0987.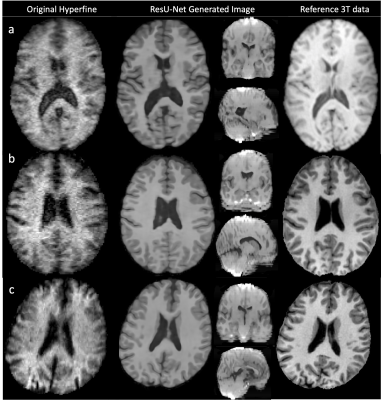 |
Refining synthetic training data improves image quality transfer
for ultra-low-field structural brain MRI
Lisa Ronan1,
Sean Deoni2,
Muriel Bruchhage3,
Godwin Ogbole4,
Matteo Figini5,
Ikeoluwa Lagunju4,
Felice D'Arco6,
Helen Cross6,
Delmiro Fernandez-Reyes5,
James Cole5,
and Daniel Alexander5 1Computer Science, University College London, London, United Kingdom, 2Gates Foundation, Seattle, WA, United States, 3University of Stavanger, Stavanger, Norway, 4University of Ibadan, Ibadan, Nigeria, 5University College London, London, United Kingdom, 6Great Ormand Street Hospital for Children, London, United Kingdom Keywords: Machine Learning/Artificial Intelligence, Low-Field MRI, super-resolution, Hyperfine Computer vision methods can be used for image quality transfer (IQT) to address the poor contrast, decreased resolution and increased noise observed in MR images acquired at ultra-low magnetic fields. Current methods have been shown to produce high-quality synthetic outputs for low-field (~0.5T) but not ultra-low-field (~0.05T) field images. Moreover, these methods do not adapt well to the presence of abnormal morphology (e.g. lesions). Here we introduce a new approach to ultra-low field IQT that improves on previous methods and is adaptive to the presence of synthetic lesions. |
| 16:57 |
0988.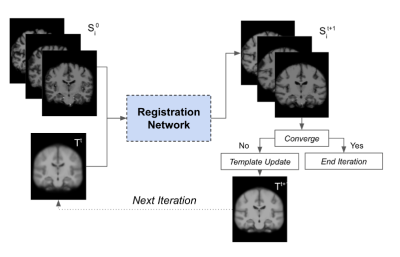 |
Constructing age-specific MRI brain templates based on a uniform
healthy population across life span with transformer
Chenghao Zhang1,
Mikhail Morgan1,
Yanting Yang1,
Ye Tian1,
and Jia Guo1
1Columbia University, New York, NY, United States Keywords: Machine Learning/Artificial Intelligence, Brain Image registration is currently employed to assist with diagnostic tasks such as neurodegenerative disease diagnosis. Aging also affects the brain, and our knowledge of age-related brain diseases is currently encumbered by age-induced bias; understanding the mechanisms of neurodegenerative diseases requires a deeper understanding of aging. Thus we have devised a pipeline for generating age-specific templates using a novel deep-learning algorithm detailed in a separate report. Our results show qualitative changes in brain morphology across age groups, and tissue segmentation was performed on each template to calculate volumetric changes in brain matter across time. |
17:05 |
0989.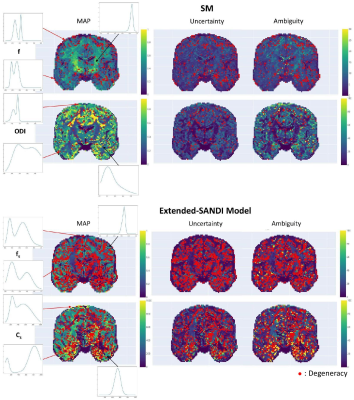 |
µGUIDE: a framework for microstructure imaging via generalized
uncertainty-driven inference using deep learning
Maëliss Jallais1,2 and
Marco Palombo1,2
1Cardiff University Brain Research Imaging Centre, Cardiff University, Cardiff, United Kingdom, 2School of Computer Science and Informatics, Cardiff University, Cardiff, United Kingdom Keywords: Signal Modeling, Microstructure, dMRI This work proposes µGUIDE: a general Bayesian framework to estimate posterior distributions of tissue microstructure parameters from any given biophysical model or MRI signal representation, with exemplar demonstration in diffusion-weighted MRI. Harnessing a new deep learning architecture for automatic signal feature selection combined with simulation-based inference and efficient sampling of the posterior distributions, µGUIDE bypasses the high computational and time cost of conventional Bayesian approaches and does not rely on acquisition constraints to define model-specific summary statistics. The obtained posterior distributions allow to highlight degeneracies present in the model definition and quantify the uncertainty and ambiguity of the estimated parameters. |
| 17:13 |
0990.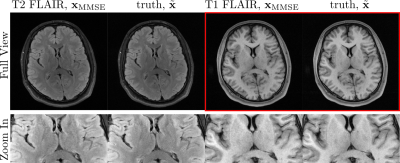 |
MRI reconstruction via Data-Driven Markov Chains with Joint
Uncertainty Estimation: Extended Analysis
Guanxiong Luo1,
Martin Heide1,
Moritz Blumenthal1,
and Martin Uecker1,2,3,4
1University Medical Center Göttingen, Göttingen, Germany, 2Institute of Biomedical Imaging, Graz University of Technology, Graz, Austria, 3German Centre for Cardiovascular Research (DZHK), Partner Site Göttingen, Göttingen, Germany, 4Cluster of Excellence "Multiscale Bioimaging: from Molecular Machines to Networks of Excitable Cells'' (MBExC), University of Göttingen, Göttingen, Germany Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction The application of generative models in MRI reconstruction is shifting researchers’ attention from the unrolled reconstruction networks to the probabilistically tractable iterations and permits an unsupervised fashion for medical image reconstruction. |
| 17:21 |
0991.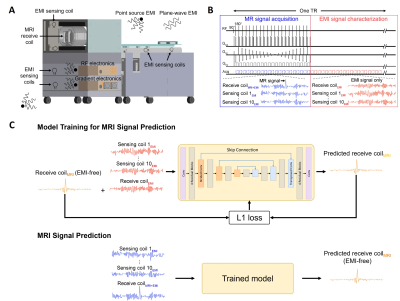 |
Robust Electromagnetic Interference (EMI) Elimination for RF
Shielding-Free MRI via Active EMI Sensing and Deep Learning MRI
Signal Prediction
Yujiao Zhao1,2,
Jiahao Hu1,2,
Vick Lau1,2,
Linfang Xiao1,2,
Alex T. L. Leong1,2,
and Ed X. Wu1,2 1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China Keywords: System Imperfections: Measurement & Correction, New Signal Preparation Schemes MRI scans are commonly performed inside a fully-enclosed RF shielding room, posing stringent installation requirement and unnecessary patient discomfort. This study develops a strategy of active EMI sensing and deep learning MR signal prediction using residual U-Net for RF shielding-free MRI. We implemented it on an ultra-low-field 0.055T head MRI scanner. Our experimental results demonstrated that this strategy could directly and accurately predict EMI-free MRI signals from the signals acquired by MRI receive coil and EMI sensing coils. It worked robustly with strong and dynamically varying EMI sources, yielding significantly improved brain image quality. |
17:29 |
0992.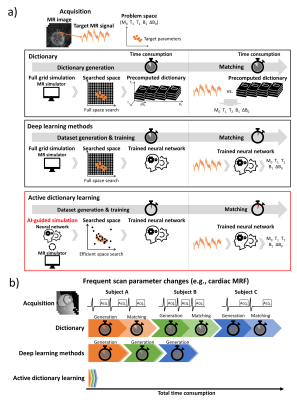 |
Active dictionary learning: fast and adaptive parameter mapping
for dynamic MRI
Hongjun An1,
Jiye Kim1,
and Jongho Lee1
1Seoul National University, Seoul, Korea, Republic of Keywords: Machine Learning/Artificial Intelligence, MR Fingerprinting
A new parameter mapping method, active dictionary
learning, for dynamic MRI is proposed. This method
trains a neural network adaptively by AI-guided MR
signal simulation. For an MRF sequence with M0,
T1,
T2,
B1,
and ΔB0,
our method successfully estimates the parameters much
faster than conventional methods (ours: 30 min for whole
process; dictionary methods: 6 hours for generation, 36
hours or 3 hours for matching). AI-guided active
dictionary learning enables adaptive quantification of
out-of-range parameters and efficient computation,
suggesting the usefulness of the method not only in
dynamic imaging but also in applications where
adaptation to parameters is necessary.
|
| 17:37 |
0993.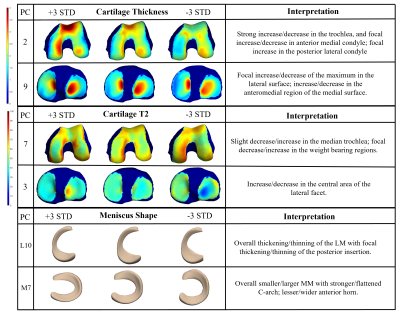 |
Quantitative MRI Interpretable 100D Feature Space of Knee
Osteoarthritis
Gabrielle Hoyer1,
Kenneth Gao1,
Jinhee Lee1,
Johanna Luitjens2,
Felix Gassert2,
Sharmila Majumdar2,
and Valentina Pedoia2 1Department of Radiology and Biomedical Imaging, UCSF; University of California Berkeley–University of California San Francisco Graduate Program in Bioengineering, University of California, San Francisco; University of California Berkeley, San Francisco, CA, United States, 2Department of Radiology and Biomedical Imaging, UCSF, University of California, San Francisco, San Francisco, CA, United States Keywords: Osteoarthritis, MSK, statistical shape modeling, cartilage thickness, cartilage T2, meniscus While the Osteoarthritis Initiative data has been explored in independent studies, to our knowledge, never has such a comprehensive analysis been completed, investigating morphology of femur, patella, tibia, menisci, for biomarkers: cartilage thickness, cartilage T2, bone shape, and meniscus shape. With 4,796 subjects from the OAI, this study utilizes an automatic statistical shape modeling technique to create a 100D interpretable space of biomarker-tissue PC modes, which can be disseminated using an innovative opensource app. In combination with Image Twin and Clinical Twin analyses, variations in morphology, as well as clinical metadata, were identified as being significantly associated with osteoarthritis incidence. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.