Power Pitch
Pitch: ML/AI Showcase
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| 13:30 |
0939.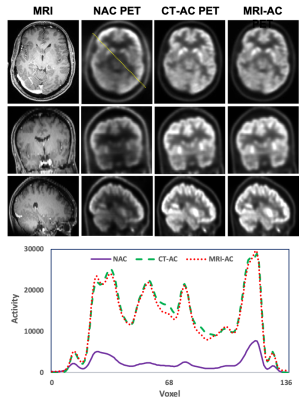 |
MRI-based Direct PET Attenuation Correction Using
Cross-modality Attention Network for Combined PET/MRI
Xiaofeng Yang1,
Yang Lei1,
Tonghe Wang2,
Sagar Mandava3,
Tian Liu4,
Jonathon Nye1,
and Hui Mao1
1Emory University, Atlanta, GA, United States, 2Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3GE Healthcare, Atlanta, GA, United States, 4Icahn School of Medicine at Mount Sinai, New York, NY, United States Keywords: PET/MR, Machine Learning/Artificial Intelligence This study aims to develop an efficient and clinically applicable deep-learning method using routine T1-weighted MRI and non-AC PET to directly synthesize high-quality attenuation-corrected PET without the need of an attenuation coefficient map for simultaneous PET/MRI. |
| 13:30 |
0940.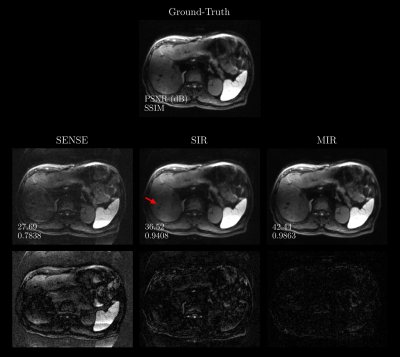 |
Joint Reconstruction of Image Repetitions in DWI using
Cross-Instance Attention
Fasil Gadjimuradov1,2,
Laura Pfaff1,
Thomas Benkert2,
Marcel Dominik Nickel2,
and Andreas Maier1
1Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, Transformer, Multiple Instance Learning Despite its proven clinical value, Diffusion-weighted Imaging (DWI) suffers from several technical limitations associated with prolonged echo trains in single-shot sequences. Parallel Imaging with sufficiently high under-sampling enabled by Deep Learning-based reconstruction may mitigate these problems. Newly emerged architectures relying on transformers demonstrated high performance in this context. This work aims at developing a transformer-based reconstruction method tailored to DWI by utilizing the availability of multiple image instances for a given slice. Redundancies are exploited by jointly reconstructing images using attention mechanisms which are performed across the set of instances. Benefits over reconstructing images separately from each other are demonstrated. |
| 13:30 |
0941.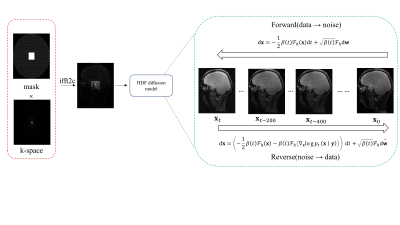 |
Diffusion generative prior-based highly accelerated MR T1ρ
mapping
Kangping Wang1,
Chentao Cao1,
Zhuoxu Cui1,
Yuanyuan Liu1,
Hairong Zheng1,
Dong Liang1,
and Yanjie Zhu1
1Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences, Shenzhen, China Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction Diffusion-based generative models have been applied to solve the inverse problem of MR reconstruction and show impressive results. However, the diffusion model requires many iterations to produce high-quality samples, prolonging the reconstruction time. It also may lead to stochastic differential equation (SDE) sequence divergence in MR reconstruction and degrades the reconstruction quality. We proposed a new SDE for diffusion-based MR reconstruction that focuses on the diffusion process in high-frequency of k-space to improve reconstruction robustness and reduce the iterations. We applied the proposed method in MR T1ρ mapping, showing that it can achieve a high acceleration of 14X. |
| 13:30 |
0942.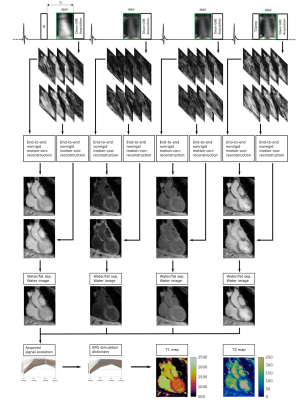 |
Deep learning-based non-rigid motion-corrected
reconstruction for whole-heart multi-dimensional joint T1/T2
imaging
Lina Felsner1,
Carlos Velasco1,
Haikun Qi2,
Karl P. Kunze1,3,
Radhouene Neji1,3,
René M. Botnar1,
and Claudia Prieto1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 3MR Research Collaborations, Siemens Healthcare Limited, Camberley, United Kingdom Keywords: Machine Learning/Artificial Intelligence, Motion Correction Myocardial T1 and T2 mapping play an important role in the assessment of cardiovascular disease. 3D whole-heart joint T1/T2 water/fat mapping approaches have been recently proposed, however they require long reconstruction times. Recently a Machine learning based reconstruction was proposed for joint motion correction and motion corrected image reconstruction of undersampled free-breathing single contrast 3D coronary MR angiography. Here, we extend this approach for non-rigid motion-corrected reconstructions for multi-contrast data for joint T1/T2 mapping. The proposed approach achieves good agreement with reference techniques and comparable image quality to state-of-the-art methods albeit in 1200 times shorter reconstruction times. |
| 13:30 |
0943.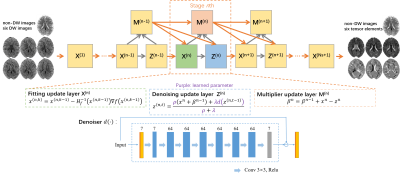 |
Diffusion Tensor Estimation Using Model-Based Deep Learning
Jialong Li1,
Qiqi Lu1,
Yanqiu Feng1,
and Xinyuan Zhang1
1Department of Biomedical Engneering, Southern Medical University, Guangzhou, China Keywords: Machine Learning/Artificial Intelligence, Diffusion Tensor Imaging Diffusion tensor imaging (DTI) is widely used in clinical applications and neuroscience. Its practical utility is limited by the need for multiple scans. Here, we integrate deep learning and model-based optimization methods to estimate diffusion tensor using only one non-diffusion-weighted images and six diffusion-weighted images. The data fidelity term is the weighted linear least squares fitting (WLLS) and the regularization term is Regularization by Denoising (RED). The Alternating Direction Method of Multiplier (ADMM) is adopted to iteratively optimize the model. Experiment results demonstrate that the proposed model-based strategy has great potential to improve the accuracy of diffusion tensor estimation. |
| 13:30 |
0944.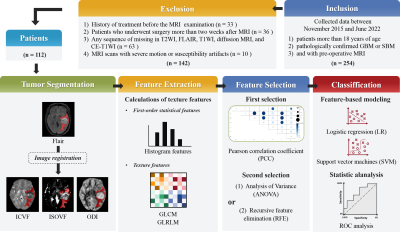 |
Neurite orientation dispersion and density imaging-based
texture features in differentiating glioblastoma from
solitary brain metastasis
Guohua Zhao1,
Mengyang He2,
Yizhou Su3,
Yusong Lin3,
Eryuan Gao1,
Jie Bai1,
Xiaoyue Ma1,
Huiting Zhang4,
Xu Yan4,
Guang Yang5,
and Jingliang Cheng1
1The Department of Magnetic Resonance Imaging, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 2School of Cyber Science and Engineering, Zhengzhou University, Zhengzhou, China, 3Collaborative Innovation Center for Internet Healthcare, Zhengzhou, China, 4MR Scientific Marketing, Siemens Healthcare, Shanghai, China, 5Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China Keywords: Machine Learning/Artificial Intelligence, Tumor Preoperative differentiation between glioblastomas (GBM) and solitary brain metastases (SBM) would aid in appropriate treatment planning and follow-up. Neurite orientation dispersion and density imaging (NODDI) can identify diverse tissue components within tumors. Texture analysis can be used to extract and quantify these tissue inhomogeneities. In this study, we extracted texture features using NODDI and validated the NODDI metric map models and a combination model to discriminate between GBM and SBM. Finally, the combined NODDI model achieved the best discriminative power. Texture analysis based on NODDI has great potential for distinguishing between GBM and SBM. |
| 13:30 |
0945.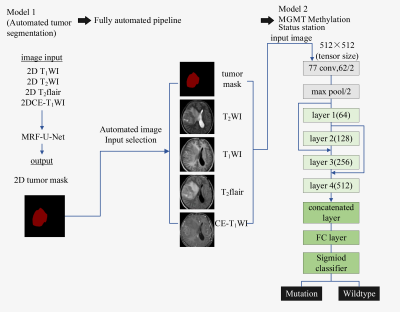 |
Prediction of MGMT Methylation Status of Gliomas Using
Pre-operative MR Images: a Fully Automatic Convolutional
Neural Networks Based Approach
Xiaohua Chen1,
Zhiqiang Chen2,
Zhuo Wang1,
Shaoru Zhang1,
Yunshu Zhou1,
Shili Liu1,
Ruodi Zhang1,
Yuhui Xiong3,
and Aijun Wang4
1Clinical medicine school of Ningxia Medical University, Yinchuan, China, 2Department of Radiology ,the First Hospital Affiliated to Hainan Medical College, Haikou, China, 3GE Healthcare MR Research, Beijing, China, 4Department of Radiology, General Hospital of Ningxia Medical University, Yinchuan, China Keywords: Machine Learning/Artificial Intelligence, Brain This study aims to propose a fully automatic approach based on convolutional neural networks (CNNs) to predict the O6-Methylguanine-DNA-methyltransferase (MGMT) methylation status of gliomas using conventional pre-operative MR images. It was shown that the Markov Random Field-U-Net network can accurately segment the tumor region, and the improved 34-layer Resnet network can predict the MGMT methylation status effectively. This model has the potential to be a practical tool for the non-invasive characterization of gliomas to help the individualized treatment planning. |
| 13:30 |
0946.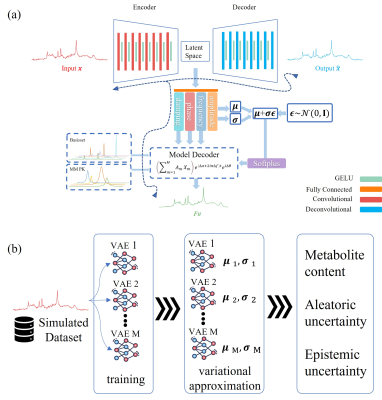 |
Physics-Informed Deep Learning Approach to Quantifying
Magnetic Resonance Spectroscopy Data with Simultaneous
Uncertainty Estimation
Amirmohammad Shamaei1,2 and
Rudy Rizzo3,4
1Institute of Scientific Instruments of the Czech Academy of Sciences Research institute in Brno, Brno, Czech Republic, 2Department of Biomedical Engineering, Brno University of Technology, Brno, Czech Republic, 3Magnetic Resonance Methodology, Institute of Diagnostic and Interventional Neuroradiology, University of Bern, Bern, Switzerland, Bern, Switzerland, 4Translational Imaging Center, sitem-insel, Bern, Switzerland Keywords: Machine Learning/Artificial Intelligence, Spectroscopy, deep learning, machine learning, convolutional neural network, metabolite quantification, Bayesian neural network, variational autoencoder, epistemic uncertainty, aleatoric uncertainty While deep learning (DL)-based approaches have been employed to quantify MRS signals, the interpretability of deep-model results and assessing what a DL model knows is a crucial component of DL-based approaches. We present a physics-informed DL-based algorithm for MRS data quantification with simultaneous uncertainty estimation, which uses the advantages of linear combination model fitting and the capabilities of ensembles of variational autoencoders. We acknowledge the need for further investigation with in-vivo datasets and other approaches. Furthermore, a more thorough analysis that includes scores that take estimation variation and uncertainty for both the proposed DL technique and traditional model fitting is required. |
| 13:30 |
0947.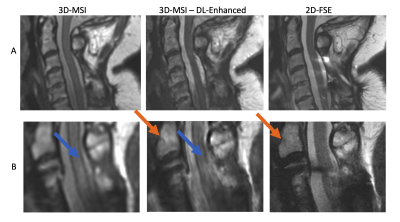 |
Deep-Learning Enhancement of 3D-MSI using Conventional
Images as Training Targets
Kevin Koch1,
Nikolai Mickevicius1,
Robin Ausman1,
and Andrew S Nencka1
1Radiology, Medical College of Wisconsin, Milwaukee, WI, United States Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction We present a feasibility study exploring the use deep learning (DL) methods to enhance the image quality of isotropic 3D-multi-spectral metal artifact suppressed images. Conventional high resolution 2D fast/turbo spin echo images are used as network training labels by masking regions corrupted by metal artifacts. A pilot set of 3 cases deploying the presented concept to T2-weighted imaging of the instrumented spine are analyzed. The quantitative results of this analysis demonstrate improved spinal cord contrast, image resolution, and general agreement with 2D-FSE images when the DL enhancement model is inferred on the complete 3D-MSI datasets. |
| 13:30 |
0948.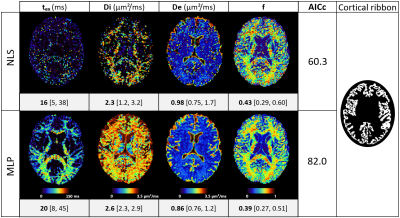 |
Optimizing the NEXI acquisition protocol for quantifying
human gray matter microstructure on a clinical MRI scanner
using Explainable AI
Quentin Uhl1,
Tommaso Pavan1,
Thorsten Feiweier2,
Erick Jorge Canales-Rodríguez3,
and Ileana Jelescu1
1Dept. of Radiology, Lausanne University Hospital (CHUV), Lausanne, Switzerland, 2Siemens Healthcare GmbH, Erlangen, Germany, 3Signal Processing Lab 5 (LTS5), École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland Keywords: Microstructure, Modelling, Acquisition Protocol We optimized the acquisition protocol for parameter estimation of the NEXI model, suited to characterize gray matter microstructure, using concepts from Explainable AI. The improvement over a “naïve” protocol was only marginal. The limit on NEXI parameter estimation precision and accuracy is largely driven by the model and the type of measurements available (linear diffusion encoding, in a combination of (b,t) pairs) and is likely already reached as can be estimated from CRLB. The silver lining is that a clinical acquisition protocol feasible on a 3T system with 80 mT/m gradients yields reasonable NEXI microstructure maps in the human brain. |
| 13:30 |
0949.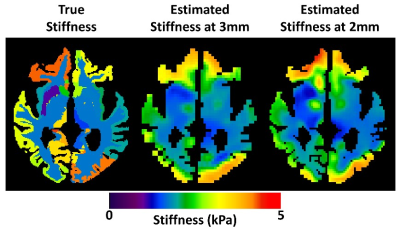 |
Impact of image resolution on brain stiffness estimation
using neural network inversion
Jonathan Trevathan1,
Jonathan Scott1,
Joshua Trzasko1,
Armando Manduca1,
John Huston1,
Richard Ehman1,
and Matthew Murphy1
1Mayo Clinic, Rochester, MN, United States Keywords: Data Acquisition, Data Acquisition, MRE,Elastrography To improve the clinical value of brain stiffness measurements, increased resolution is desired to accurately map the mechanical signatures of disease processes. However, MR elastography-based stiffness maps must be estimated by inverting the measured displacement fields. It is not well-established that increased acquisition resolution will directly translate to improved accuracy in the final mechanical property maps. In this simulation study, using two neural network inversions, we show 2-mm data outperforms 3-mm data for stiffness accuracy and precision across a range of signal-to-noise ratios. |
| 13:30 |
0950.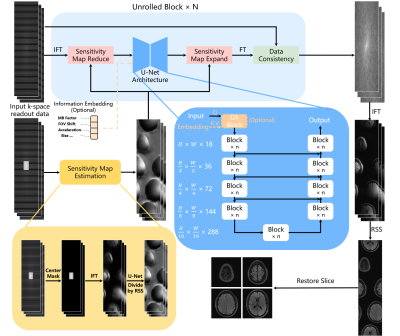 |
VarNet-based Simultaneous Multislice Reconstruction for Low
Noise Amplification and Flexible Calibration
Lifeng Mei1,
Celia.M. Dong2,
Sixing Liu1,
Shoujin Huang1,
and Mengye Lyu1
1College of Health Science and Environmental Engineering, Shenzhen Technology University, Shenzhen, China, 2University Research Facility in Behavioral and Systems Neuroscience, The Hong Kong Polytechnic University, Hong Kong, China Keywords: Image Reconstruction, Parallel Imaging In this study, we combine the readout-concatenation framework with VarNet based deep learning method to achieve SMS reconstruction of low noise amplification. It can flexibly handle arbitrary slice aliasing patterns with in-plane acceleration. Moreover, the modified network architecture allows coil calibration using a prescan of different contrasts, which is preferred in many scenarios. |
| 13:30 |
0951.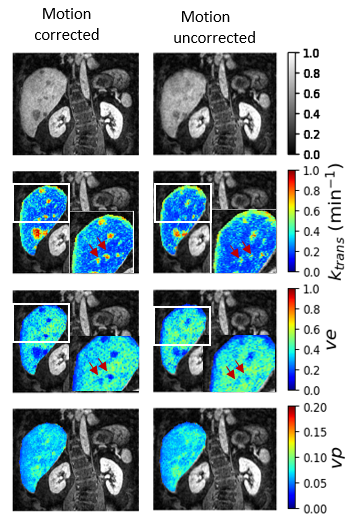 |
Deep learning for quantitative evaluation of motion
correction in dynamic contrast enhanced (DCE) MRI
Edengenet Mashilla Dejene1,2 and
Christoph Kolbitsch1
1Physikalisch - Technische Bundesanstalt (PTB), Braunschweig and Berlin, Germany, 2Charité Universitätsmedizin Berlin, Berlin, Germany Keywords: Liver, DSC & DCE Perfusion Respiratory motion can impair the accurate estimation of physiological parameters in DCE-MR of the liver. A Deep learning network is proposed to quantitatively investigate the impact of respiratory motion on the estimation of physiological parameter maps. The proposed network provides quantitative parameters for DCE-MR and uncertainty estimates for these parameters. Here we could show that the estimated epistemic uncertainty of k_trans is sensitive to motion. This could provide important information about how well motion correction worked and how reliable the obtained quantitative DCE parameters are. |
| 13:30 |
0952.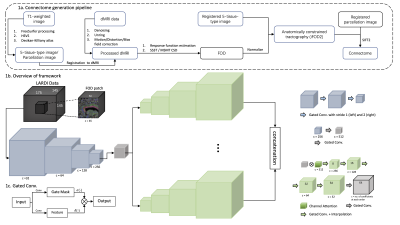 |
FOD-Net 2.0: End-to-end FOD enhancement for low angular
diffusion acquisitions using deep learning
Xinyi Wang1,2,
Zihao Tang1,2,
Mariano Cabezas2,
Arkiev D’Souza2,
Fernando Calamante2,
Dongnan Liu1,2,
Michael Barnett2,3,
Sicong Tu2,
Weidong Cai1,
and Chenyu Wang2,3
1School of Computer Science, University of Sydney, Sydney, Australia, 2Brain and Mind Centre, University of Sydney, Sydney, Australia, 3Sydney Neuroimaging Analysis Centre, University of Sydney, Sydney, Australia Keywords: Brain Connectivity, Tractography & Fibre Modelling, Fiber Orientation Distribution Modern structural brain connectome pipelines and tractography techniques heavily rely on the quality of the diffusion weighted image acquisition (angular resolution) and the subsequent estimation of the fiber orientation distributions (FODs) for each voxel. Generating reliable connectomes from low angular single-shell acquisitions in clinical scenarios remains a challenging task. This work presents an end-to-end deep learning framework to enhance FOD estimates according to multi-shell acquisitions from low angular single-shell acquisitions to guarantee high quality tractography and connectomes within acceptable time and resources. |
| 13:30 |
0953.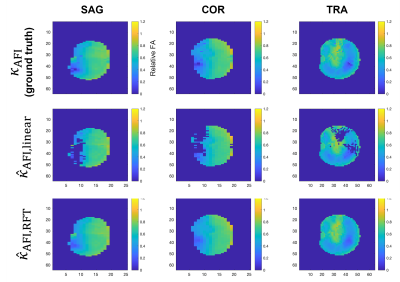 |
A Machine-Learning Approach for Saturation-Prepared Turbo
FLASH B1+ Maps Calibration
Michael Dubiner1,
Jan Sedlacik1,2,3,4,
Tom Wilkinson1,2,3,
Pip Bridgen1,2,3,
Franck Mauconduit5,
Alexis Amadon5,
Sharon Giles1,2,3,
Radhouene Neji1,3,6,
Joseph V. Hajnal1,2,3,
Shaihan J. Malik1,2,3,
and Raphael Tomi-Tricot1,2,3,6
1Biomedical Engineering Department, School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom, 2Centre for the Developing Brain, School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom, 3London Collaborative Ultra high field System (LoCUS), London, United Kingdom, 4Great Ormond Street Hospital for Children, London, United Kingdom, 5Paris-Saclay University, CEA, CNRS, BAOBAB, NeuroSpin, Gif-sur-Yvette, France, 6MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom Keywords: RF Pulse Design & Fields, High-Field MRI A fast B1+ mapping sequence such as saturation-prepared turbo FLASH (satTFL) is desirable for online pulse design at ultra-high field, but it often results in inaccuracies. Previous work performed linear fitting of the B1+ magnitude obtained with the satTFL over that of the longer but more accurate Actual Flip angle Imaging (AFI) sequence to obtain calibration parameters that would correct the satTFL B1+ on the fly for brain imaging. In this work we introduce a new machine-learning-based method that uses additional features to create a more precise and less location-dependent accuracy. |
| 13:30 |
0954.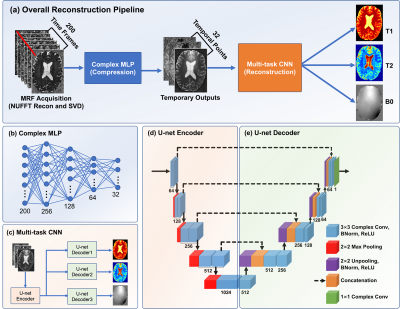 |
MRF-mixer: a self-supervised deep learning MRF framework
Yang Gao1,2,
Tianyi Ding2,
Martijn Cloos2,
and Hongfu Sun2
1Central South University, Changsha, China, 2The University of Queensland, Brisbane, Australia Keywords: Contrast Mechanisms, MR Fingerprinting, relaxometry reconstruction, B0 estimation, self-supervised deep learning Deep neural networks are increasingly employed in MRF reconstruction recently, as an alternative for conventional dictionary matching. However, most previous deep leanring MRF networks were trained based on the dictionary pairs (generated using theoretical bloch equation) or in vivo acquisitions paired with dictionary matching reconstructions (as training labels), whose reconstruction accuracy relies heavily on the dictionary construction and the number of sampling arms. In this work, we propose a self-supversied deep learning MRF method, namely MRF-Mixer, which can result in more accurate MRF reconstructions compared with dictionary matching. |
| 13:30 |
0955.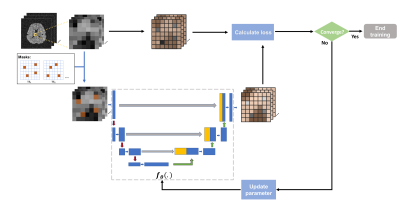 |
MR Spatiospectral Reconstruction using Plug&Play Denoiser
with Self-Supervised Training
Ruiyang Zhao1,2,
Yahang Li1,3,
Zepeng Wang1,3,
Aaron Anderson1,4,
Paul Arnold1,4,
Graham Huesmann1,4,5,
and Fan Lam1,2,3
1Beckman Institute for Advanced Science and Technology, Urbana, IL, United States, 2Department of Electrical and Computer Engineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 3Department of Bioengineering, University of illinois Urbana-Champaign, Urbana, IL, United States, 4Neuroscience Institute, Carle Foundation Hospital, Urbana, IL, United States, 5School of Molecular and Cellular Biology, University of Illinois Urbana-Champaign, Urbana, IL, United States Keywords: Spectroscopy, Machine Learning/Artificial Intelligence We introduced a data-driven denoiser trained in a self-supervised fashion as a novel spatial-temporal constraint for MRSI reconstruction. Our proposed denoiser was trained using noisy data only via the Noise2void framework that trains an interpolation network exploiting the statistical differences between spatiotemporally correlated signals and uncorrelated noise. The trained denoiser was then integrated into an iterative MRSI reconstruction formalism as a Plug-and-Play prior. An additional physics-based subspace constraint was also included into the reconstruction. Simulation and in vivo results demonstrated impressive SNR-enhancing reconstruction ability of the proposed method, with improved performance over a state-of-the-art subspace method. |
13:30 |
0956.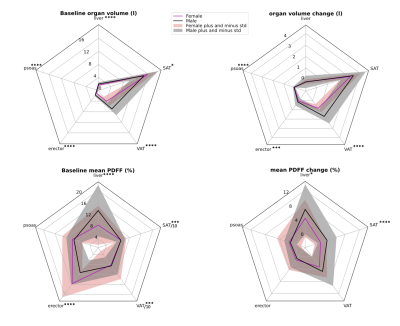 |
Assessing Sex Differences in Abdominal Fat Depots of People
with Obesity under Weight Loss using Automated Segmentation
Mingming Wu1,
Arum Somasundaram1,
Selina Rupp1,
Jessie Han1,
Stella Naebauer1,
Daniela Junker1,
Anna Reik2,
Meike Wiechert2,
Hans Hauner2,3,
Christina Holzapfel2,
and Dimitrios C. Karampinos1
1Department of Diagnostic and Interventional Radiology, School of Medicine, Technical University of Munich, School of Medicine, Technical University of Munich, Munich, Germany, 2Institute for Nutritional Medicine, School of Medicine, Technical University of Munich, Munich, Germany, 3Else Kroener-Fresenius-Center of Nutritional Medicine, School of Life Sciences, Technical University of Munich, Freising, Germany Keywords: Fat, Metabolism, Obesity A deep-learning algorithm based on the nnU-Net and using water-fat images enabled robust automatic segmentation of abdominal organs including visceral and subcutaneous adipose tissue, liver, iliopsoas and erector spinae muscle groups. Each organ's volume and fat content were examined in a weight loss study comprising 127 subjects with BMI of 30-39.9kg/m2, who followed a low caloric diet (LCD). Dixon water-fat images were acquired before and after diet. Differences in fat distribution among abdominal organs and fat content was assessed among both sexes. Differences in the changes of organ volume and fat fraction as a response to the LCD were revealed. |
| 13:30 |
0957.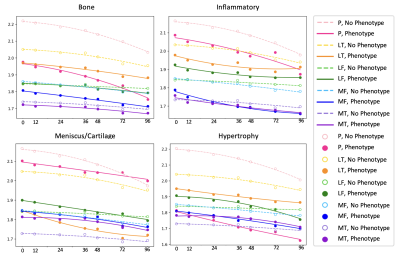 |
Variation in 8 years Cartilage Loss Trajectories Based on
Baseline Pain and Deep Learning-Assessed ROAMES
Morphological Phenotypes
Madeline Hess1,
Sharmila Majumdar1,
and Valentina Pedoia1
1University of California San Francisco, San Francisco, CA, United States Keywords: Osteoarthritis, Osteoarthritis We find that the presence or absence of ROAMES morphological phenotypes as predicted from MRI is a useful indicator of osteoarthiris progression over 8 years as measured by cartilage thinning. |
| 13:30 |
0958.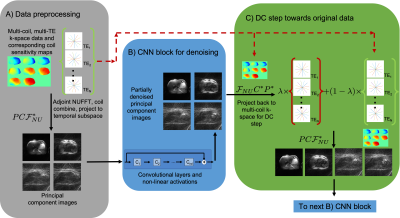 |
Deep learning reconstruction of radial T2 weighted data sets
with data consistent unrolled neural networks
Brian Toner1,
Simon Arberet2,
Fei Han3,
Mariappan Nadar2,
Vibhas Deshpande4,
Diego Martin5,
Maria Altbach6,
and Ali Bilgin7,8
1Applied Mathematics, The University of Arizona, Tucson, AZ, United States, 2Digital Technology & Innovation, Siemens Healthineers, Princeton, NJ, United States, 3Siemens Healthineers, Los Angeles, CA, United States, 4Siemens Healthineers, Austin, TX, United States, 5Radiology, Houston Methodist, Houston, TX, United States, 6Medical Imaging, The University of Arizona, Tucson, AZ, United States, 7Electrical and Computer Engineering, The University of Arizona, Tucson, AZ, United States, 8Biomedical Engineering, The University of Arizona, Tucson, AZ, United States Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction Unrolled networks with data consistency layers have been shown to be effective in reconstructing MRI data. Radial turbo spin echo sequences enable acquisition of multi-contrast k-space data, which can be used to generate multi-contrast images at different echo times together with a co-registered T2 map. In this work, we will show that the cascading unrolled network architecture is effective in reconstructing images from radial turbo spin echo data. In order to do this, data consistency layers must be implemented to be able to combine data from multi-coil acquisitions and from multiple echo times. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.