Traditional Poster
Acquisition & Analysis
ISMRM & ISMRT Annual Meeting & Exhibition • 03-08 June 2023 • Toronto, ON, Canada

| Booth # | |||
|---|---|---|---|
5254.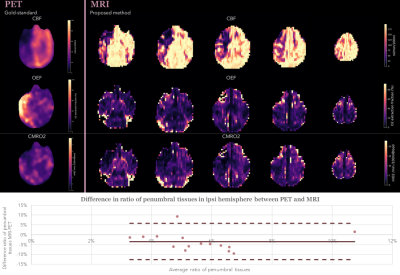 |
Translational validation of a MRI-based CMRO2 mapping method for
penumbra definition in acute ischemic stroke
Lucie Chalet1,2,
Timothé Boutelier2,
Thomas Christen3,
Justine Debatisse4,
Oceane Wateau5,
Nicolas Costes6,
Ines Merida6,
Sophie Lancelot6,7,
Christelle Leon1,
Norbert Nighoghossian1,8,
Yves Berthezene9,10,
Omer Faruk Eker9,10,
Tae-Hee Cho1,8,
Emmanuelle Canet-Soulas1,
and Laura Mechtouff1,8
1Univ Lyon, CarMeN Laboratory, INSERM, INRA, INSA Lyon, Universite Claude Bernard Lyon 1, Lyon, France, 2Olea Medical, La Ciotat, France, 3Univ. Grenoble Alpes, Inserm, U1216, Grenoble Institut Neurosciences, Grenoble, France, 4Institut des Sciences Cognitives Marc Jeannerod (ISCMJ), Unite Mixte de Recherche 5229 du Centre National de la Recherche Scientifique (CNRS), Bron, France, 5Cynbiose SAS, Marcy-L'Etoile, France, 6CERMEP - Imagerie du Vivant, Lyon, France, 7Hospices Civils of Lyon, Lyon, France, 8Stroke department, Hospices Civils of Lyon, Lyon, France, 9CREATIS, CNRS UMR-5220, INSERM U1206, Universite Lyon 1, INSA Lyon, Villeurbanne, France, 10Neuroradiology department, Hospices Civils of Lyon, Lyon, France Keywords: Data Processing, Oxygenation, Oxygen Metabolism Latest challenges in acute ischemic stroke management involve tissue-based approaches to define a tailored ischemic penumbra to each patients. Oxygen metabolism imaging offers new perspectives to tackle these challenges. A fully automated pipeline was developed providing oxygen metabolism parameters from MRI acquisitions including advanced processing methods such as motion-correction, registration and bayesian-based deconvolution. The method was tested on an ischemia-reperfusion non-human primate model with simultaneous PET-MRI acquisitions providing gold-standard comparison of oxygen parameters. Results show strong similarities in ratio of penumbral tissues detected in the ischemic hemisphere between the two modalities and opens new perspectives of improvement in parameters definition. |
||
5255.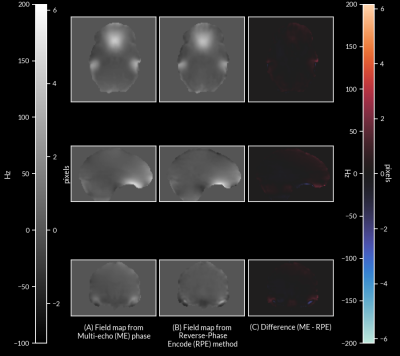 |
Framewise distortion correction in Multi-echo Echo Planar
Imaging
Andrew N. Van1,2,
M. Dylan Tisdall3,
Timothy O. Laumann4,
David F. Montez2,4,
Damien A. Fair5,6,7,
and Nico U.F. Dosenbach1,2,8,9
1Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO, United States, 2Department of Neurology, Washington University School of Medicine, St. Louis, MO, United States, 3Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 4Department of Psychiatry, Washington University School of Medicine, St. Louis, MO, United States, 5Institute of Child Development, University of Minnesota Medical School, Minneapolis, MN, United States, 6Department of Pediatrics, University of Minnesota Medical School, Minneapolis, MN, United States, 7Masonic Institute for the Developing Brain, University of Minnesota Medical School, Minneapolis, MN, United States, 8Department of Radiology, Washington University School of Medicine, St. Louis, MO, United States, 9Department of Pediatrics, Washington University School of Medicine, St. Louis, MO, United States Keywords: Data Processing, fMRI Echo Planar Imaging (EPI) suffers from off-resonance image distortion due to B0 inhomogeneities. These distortions are typically corrected by separate field map acquisitions, but only allow for sparse temporal sampling of B0 inhomogeneity. This approach prevents distortion correction if changes in the B0 field occur over time due to motion. Here, we present a method of correcting off-resonance distortion by computing a time series of field maps using phase information from multi-echo (ME) EPI data. We show that our method produces corrections comparable to the current standard of distortion correction, in addition to enabling frame-by-frame correction in functional MRI data. |
||
5256.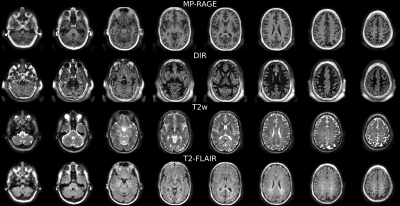 |
Multicontrast ZTE Neuroimaging with T1-Shuffling
Tobias C Wood1 and
Emil Ljungberg1,2
1King's College London, London, United Kingdom, 2Lund University, Lund, Sweden Keywords: Image Reconstruction, Image Reconstruction We show that multiple clinical contrasts can be obtained from silent Zero Echo Time scans by using preparation pulses and a physics-informed subspace "T1-shuffling" reconstruction approach. This models and removes the effect of T1-recovery during the readout segment, which would otherwise wash out the desired contrast. |
||
5257.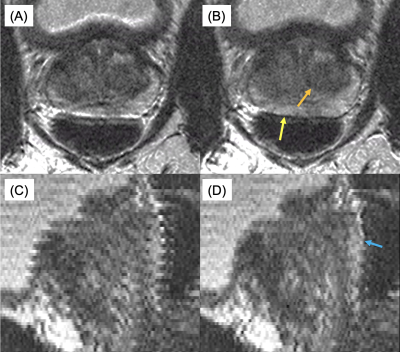 |
Navigator-Based In-Plane Motion Correction for High Resolution
2D T2-Weighted Spin-Echo Prostate MRI
Eric A. Borisch1,
Adam T Froemming1,
Roger C. Grimm1,
Daniel Reiter2,
Phillip J. Rossman1,
and Stephen J. Riederer1
1Radiology, Mayo Clinic, Rochester, MN, United States, 2St. Olaf College, Northfield, MN, United States Keywords: Motion Correction, Motion Correction The addition of ky≈0 navigator views to detect anterior-posterior prostate (peristalsis-induced) motion to the end of each echo train for a 2D T2-weighted acquisition stack with slice-profile recovery reconstruction is investigated. ROVir virtual coils are used to focus the spatial sensitivity in the phase direction of the navigator response on the small, central prostate region. Mutual information is used to register these navigators across the acquisition, with the resulting movement time course used to correct the acquired data. Results both in motion controlled phantom and human studies are discussed. |
||
5258.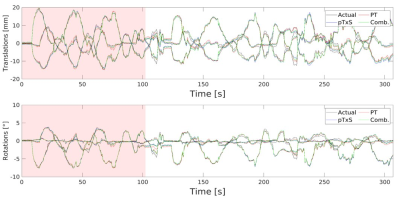 |
Pilot Tone vs pTx Scattering: A Comparison between ‘RF Sensor’
Methods for Rigid Body Motion Detection of the Brain at 7T
James L. Kent1,
Ladislav Valkovič2,3,
Iulius Dragonu4,
Mark Chiew1,5,6,
and Aaron T. Hess1
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 2Oxford Centre for Clinical Magnetic Resonance Research, John Radcliffe Hospital, University of Oxford, Oxford, United Kingdom, 3Department of Imaging Methods, Institute of Measurement Science, Slovak Academy of Sciences, Bratislava, Slovakia, 4Research & Collaborations GB&I, Siemens Healthcare Ltd, Camberley, United Kingdom, 5Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada, 6Physical Sciences, Sunnybrook Research Institute, Toronto, ON, Canada Keywords: Motion Correction, Motion Correction RF sensor methods for motion correction require no (or minimal) additional hardware, are sequence independent and have high temporal resolution. Several RF sensor-based methods for rigid-body head motion detection have been demonstrated but which method offers the most sensitivity to motion is yet unknown. We aim to compare the sensitivity of PT and pTxS methods by simultaneously measuring these signals during continuous motion, training a linear model from EPI registered images and analysing the ability to predict rigid head positions. Currently, we see little difference in their ability to predict rigid head motion but further investigation is needed. |
||
5259. |
Towards a standardized file format for basis spectra and fitting
results for linear-combination modeling
Helge J. Zöllner1,2,
Kelley M. Swanberg3,
John LaMaster4,
Antonia Kaiser5,
Jamie Near6,
Candace Fleischer7,8,
Brian J. Soher9,
William T. Clarke10,
and Georg Oeltzschner1,2
1The Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Kennedy Krieger Institute, F. M. Kirby Research Center for Functional Brain Imaging, Baltimore, MD, United States, 3Department of Biomedical Engineering, Columbia University Fu Foundation School of Engineering and Applied Science, New York, NY, United States, 4Faculty of Computer Science, Technische Universität München, München, Germany, 5Department of Radiology and Nuclear Medicine, Amsterdam Neuroscience, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands, 6Physical Sciences Platform, Sunnybrook Research Institute, Toronto, ON, Canada, 7Department of Radiology and Imaging Sciences, Emory University School of Medicine, Atlanta, GA, United States, 8Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, United States, 9Center for Advanced MR Development, Department of Radiology, Duke University Medical Center, Durham, NC, United States, 10Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom Keywords: Data Analysis, Spectroscopy, open source We propose a standardized format for storing basis sets and linear-combination modeling results. Conventions, guidelines, and conversion tools are currently developed in collaboration with the MRS community. This standardization effort will further improve data sharing and reproducibility, allowing for easier integration of MRS into larger neuroimaging frameworks. |
||
5260. |
Entropy informs about uncertainty in probabilistic brain tissue
segmentation
Agnieszka Sierhej1,2,
Matt G Hall1,2,
Nadia AS Smith2,
and Chris A Clark1
1University College London, London, United Kingdom, 2National Physical Laboratory, London, United Kingdom Keywords: Segmentation, Brain, Uncertainty Segmentation is crucial in analysing volumes of brain tissue compartments in studies of healthy development and pathological changes. Probabilistic segmentation is widely used, but the choice of threshold on probability maps has large effects on the resulting tissue volumes, and there is no consensus on optimal choice of such threshold. This work presents a way to minimise this effect by quantifying the uncertainty using Shannon entropy. Entropy map contains information about uncertainties in each individual voxel and this can be used to inform about uncertainty in MRI-based tissue segmentation. |
||
5261.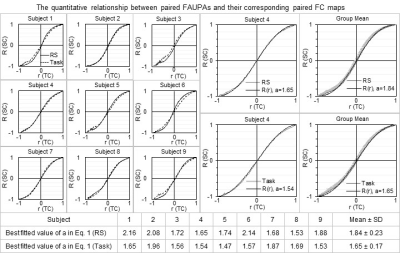 |
The relationship of brain areal activity with the entire brain’s
activity
Jie Huang1
1Michigan State University, East Lansing, MI, United States Keywords: Data Analysis, fMRI The relationship between the activity of brain areas and that of the entire brain remains unknown. The temporal correlation of the BOLD time signal of an area with that of every point in the brain yields a full spatial map that characterizes the entire brain’s functional co-activity (FC) relative to that area’s activity. Analyzing the temporal correlation of the signal time courses of two areas and the spatial correlation of their corresponding two FC maps for all pairwise areas revealed a quantitative relationship between the activity of brain areas and that of the entire brain. |
||
5262.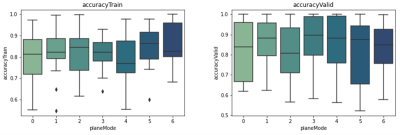 |
The impact of data normalization and plane view in deep learning
for classifying multiple sclerosis subtypes using MRI scans.
Mahshid Soleymani1,
Yunyan Zhang2,
and Mariana Bento2
1Biomedical Engineering, University of Calgary, Calgary, AB, Canada, 2University of Calgary, Calgary, AB, Canada Keywords: Machine Learning/Artificial Intelligence, Data Processing Disease activity varies between patients with multiple sclerosis (MS), and patients who have a greater risk of developing a progressive course require more aggressive therapies earlier. However, differentiating disease severity is challenging using conventional methods as the disease often progresses silently. By taking advantage of one of the most advanced quantitative methods, convolutional neural networks, we aim to develop a new deep learning model to differentiate two common MS subtypes: relapsing-remitting course from secondary progressive phenotype. This study focuses on varying image pre-processing techniques and using different data views using conventional brain MRI. |
||
5263.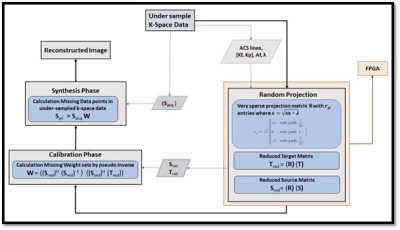 |
High-Performance FPGA-based Accelerator using Random Projection
for pMRI method
Nimra Naeem 1,
Omair Inam 2,
Abdul Basit 2,
Hammad Omer2,
and Tayaba Gul2
1Electrical and Computer Engineering, COMSATS University Islamabad, Islamabad, Pakistan, 2COMSATS University Islamabad, Islamabad, Pakistan Keywords: Parallel Imaging, Cardiovascular The random projection method reduces the dimensionality of the data to provide attractive computational advantages in the collection and processing of high-dimensional signals. In literature, it has been successfully applied for the pMRI method i.e., GRAPPA. This paper introduces a very sparse random projection matrix with the same statistical efficiency as dense matrix. The proposed method is implemented on FPGA (working with on-chip processor) device to speed up the GRAPPA reconstruction process. The reconstruction results for in vivo 30 channel human cardiac data set show 6x speed up with little loss in accuracy. |
||
5264.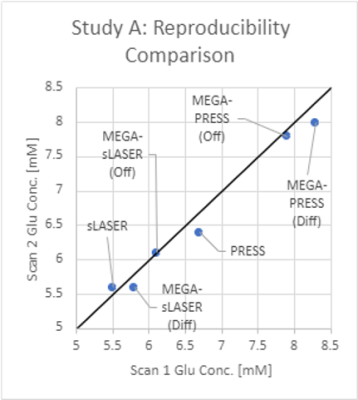 |
Reliable Glutamate Measurements at 3T: Which Sequence should I
Choose?
Philip F. Durham1,
Humberto Monsivais1,
Gianna Nossa1,
Daniel Foti2,
Xiaopeng Zhou3,
and Ulrike Dydak1,4
1College of Health and Human Sciences, Purdue University, West Lafayette, IN, United States, 2Department of Psychological Sciences, Purdue University, West Lafayette, IN, United States, 3Purdue Life Sciences MRI Facility, Purdue University, West Lafayette, IN, United States, 4Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, United States Keywords: Data Acquisition, Brain, Glutamate, MRS This meta-analysis discusses the intra- and inter-subject variability of glutamate measurements in MRS, comparing widely used sequences on clinical scanners (PRESS, sLASER, MEGA-PRESS, MEGA-sLASER, HERMES). To minimize variance in glutamate measurements, the authors recommend sLASER over PRESS, non-edited over edited MRS, and OFF over DIFF spectra. |
||
5265.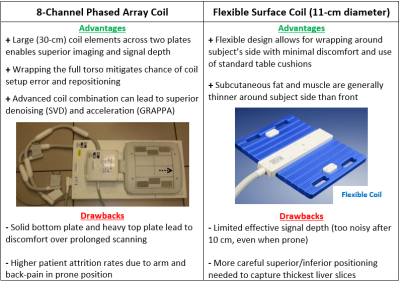 |
Optimizing a Clinically Feasible 31P Liver MRSI Protocol at 3T
Brian Bozymski1,
Nathan Ooms1,
and Ulrike Dydak1,2
1School of Health Sciences, Purdue University, West Lafayette, IN, United States, 2Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, United States Keywords: Data Acquisition, Liver, Fatty Liver Disease Optimizing a clinically feasible 31P liver MRSI protocol on a 3T SIEMENS system through the lens of a diffuse fatty liver disease study. Protocol practicality is driven primarily by scan length, patient comfort, and data quality. Voluntary scanning subjects, ranging from healthy to overweight, are sourced from the university population. Both a dual-tuned, 8-channel 1H/31P phased array coil (wrapped around the whole torso) and an 11-cm-diameter flexible surface coil are available for use. Techniques, testing results, and relative advantages of either are discussed in detail, as well as the implications for each choice in a clinical setting. |
||
5266. |
Influence of Gradient Polarity in Magnetic Resonance
Spectroscopy at 3T using HERMES
Antonia Susnjar1,
Antonia Susnjar2,
Gianna Nossa3,
and Ulrike Dydak3,4
1Biomedical Engineering, Purdue University, West Lafayette, IN, United States, 2Purdue University, West Lafayette, IN, United States, 3School of Health Sciences, Purdue University, West Lafayette, IN, United States, 4Department of Radiology and Imaging Sciences, Indiana School of Medicine, Indianapolis, IN, United States Keywords: Data Acquisition, Data Acquisition To overcome acquisition downfalls of HERMES edited magnetic resonance spectroscopy at 3T, we have conducted critical optimization for four clinically relevant brain regions in neurological disorders by finding the optimal voxel rotation that affects gradient order to enhance data quality and reproducibility. |
||
 |
5267.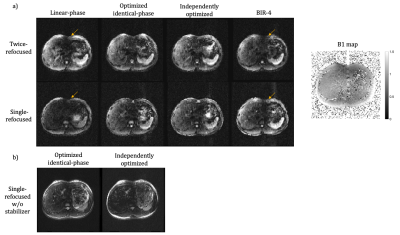 |
Optimized B0 and B1+ insensitive RF pulses for non-selective T2
and diffusion preparation
Xuetong Zhou1,2,
Brian A. Hargreaves1,2,3,
and Philip K. Lee2
1Department of Bioengineering, Stanford University, Stanford, CA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States, 3Department of Electrical Engineering, Stanford University, Stanford, CA, United States Keywords: New Signal Preparation Schemes, Data Acquisition The performance of conventional T2 or diffusion preparation using linear phase pulses or adiabatic pulses is limited by the B1+ and B0 field inhomogeneities, particularly relevant in body imaging. In this work, we proposed a numerical optimization strategy to design RF pulse pairs for non-selective T2 or diffusion preparation that perform robustly over a wide range of B1+ and B0 variations. Phantom and in vivo experiments compared the performance of preparation using the optimized pulses to that of linear-phase and adiabatic pulses. |
|
5268.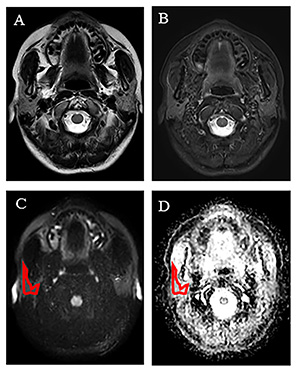 |
Radiomics analysis of apparent diffusion coefficient maps of
parotid gland to diagnose morphologically normal Sjogren’s
syndrome
Chen Chu1,
Jie Meng1,
Huayong Zhang2,
Qianqian Feng1,
Weibo Chen3,
Jian He1,
Lingyun Sun2,
and Zhengyang Zhou1
1Department of Radiology, Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, China, 2Department of Rheumatology, Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, China, 3Philips Healthcare, Shanghai, China, Shanghai, China Keywords: Radiomics, Diffusion/other diffusion imaging techniques The study explored more promising radiomics extracted from apparent diffusion coefficient maps for diagnosing Sjogren’s syndrome (SS) without head&neck MR morphology changes. A total of 119 consecutive SS participants and 95 healthy volunteers were prospectively analyzed by 3.0 T MR including diffusion weighted imaging. Forty-five radiomic parameters were selected and twenty-two radiomic parameters showed significant difference between SS and controls, in which 11 parameters had an area under the ROC curve (AUC) greater than 0.700. The SVM classification model differentiated SS from healthy controls with an AUC of 0.932 and 0.911 in the training and testing sets, respectively. |
||
5269.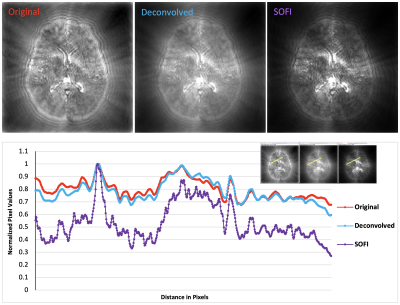 |
Super-Resolution Magnetic Resonance Fingerprinting
Judith Mizrachi1,
Marco Barbieri1,
Akshay Chaudhari1,
and Garry Gold1
1Radiology, Stanford University, Stanford, CA, United States Keywords: Hybrid & Novel Systems Technology, Data Processing, super-resolution, high resolution, Fingerprinting We introduce a novel approach, Super-Resolution Magnetic Resonance Fingerprinting (SR-MRF), which is a combination of an optical super-resolution microscopy method, Super-resolution Optical Fluctuation Imaging (SOFI) and MR-Fingerprinting. Our preliminary results in simulated MRF data demonstrate both 2-fold resolution improvement, as demonstrated by point spread function (PSF) analysis, and tissue-type segmentation. This technique represents an original crossover between the fields of MR-imaging and super-resolution microscopy, and lays the groundwork for other such techniques to follow. |
||
5270.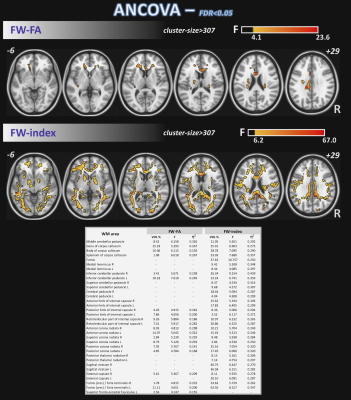 |
Neuroimaging Correlates of Functional Motor Changes in
Cognitively Impaired Cohorts
Maurizio Bergamino1,
Elizabeth G Keeling1,2,
Sydney Y Schaefer2,
Anna Burke1,
George Prigatano1,
and Ashley M Stokes1
1Barrow Neurological Institute, Phoenix, AZ, United States, 2Arizona State University, Tempe, AZ, United States Keywords: Data Analysis, Diffusion/other diffusion imaging techniques The objective of this study was to assess white matter microstructural differences between groups of healthy controls (HC), subjective memory complaints (SMC), and cognitive impairment (CI) through multi-shell free-water (FW) corrected DTI (FW-DTI) metrics and by the peak width of skeletonized mean diffusivity (PSMD). Additionally, brain correlations at the voxel level between a functional motor measure (two-bean transfer) and DTI metrics/PSMD were explored. |
||
5271.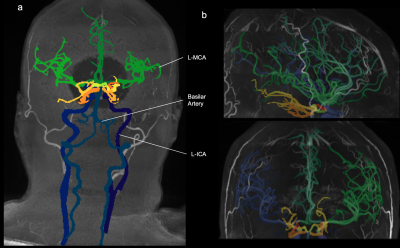 |
The Multi-Class Segmentation of the Human Cerebral Vasculature
in TOF-MRA: A Supervised Deep Learning Approach
Karim Fathy1,
Felix Dumais2,
Samantha Côté 1,
Blaise Frederick3,
and Kevin Whittingstall4
1Departement of Biomedical Imaging and Radiation Science, Universite de Sherbrooke, Sherbrooke, QC, Canada, 2Departement of Computer Science, Universite de Sherbrooke, Sherbrooke, QC, Canada, 3Departement of Psychiatry, Harvard Medical School, Belmont, MA, United States, 4Departement of Radiology, Universite de Sherbrooke, Sherbrooke, QC, Canada Keywords: Machine Learning/Artificial Intelligence, Neuro, Software Tool Currently, there is no accurate fully automated multi-class method to segment the whole cerebral arterial tree in time-of-flight magnetic resonance angiography (TOF-MRA). We developed an artificial intelligence based software tool to identify cerebral arteries in TOF-MRAs. We trained a neural network on a TOF-MRA dataset and labeled the cerebral arterial tree using different image processing techniques. Our software tool is fast and reliable, with no human intervention, and allows for the conduction of large-scale TOF-MRA studies while being versatile in segmenting a diverse set of TOF-MRAs. |
||
5272.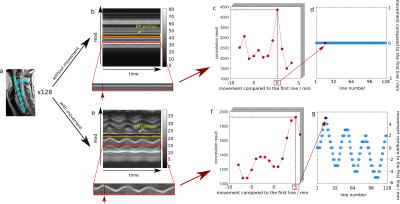 |
Detecting Spine Movements with a Navigator Echo for Motion
Correction of Spinal Cord fMRI
Ying Chu1 and
Jürgen Finsterbusch1
1Department of Systems Neuroscience, University Medical Center Hamburg-Eppendorf, Hamburg, Germany Keywords: Motion Correction, fMRI Motion correction is crucial for functional neuroimaging, however, not straightforward for applications in the cervical spinal cord because its shape, size, and inner structure barely vary along the cord’s axis. Previous spinal cord fMRI studies involved motion correction in this direction based on the vertebral disks visible in the fMRI acquisitions but for transverse slices, this approach suffers from a low spatial resolution (slice thickness typically 3.5mm or more). Here, a columnar navigator positioned along the spine is used to detect spine movements with a high spatial resolution that could be used for motion correction of spinal cord fMRI. |
||
5273.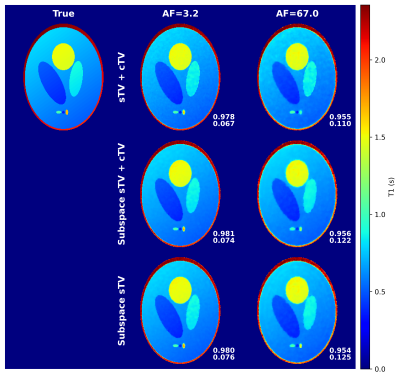 |
Principal component subspace reconstruction for compressed
sensing of radial quantitative MRI.
Antti Paajanen1,
Olli Nykänen1,
Ville Kolehmainen1,
and Mikko J. Nissi1
1Department of Applied Physics, University of Eastern Finland, Kuopio, Finland Keywords: Sparse & Low-Rank Models, Quantitative Imaging, Image reconstruction Quantitative MRI offers unique opportunities for compressed sensing reconstructions because the signal evolution is known. Here we compare a principal component subspace reconstruction to a standard total-variation regularized compressed sensing approach with a 3-D radial variable flip angle simulation data. The simulation data allows us to measure only the effect of the chosen reconstruction. The subspace approach consistently yields similar or better image quality and does not seem to require contrast dimension regularization to achieve it. |
||
5274.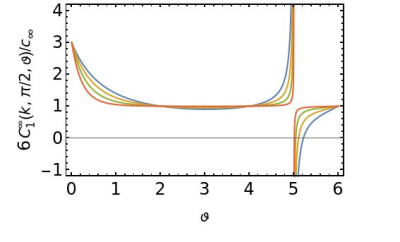 |
Apparent diffusion coefficient in the low-frequency limit when
measured with oscillating-gradient spin-echo diffusion-weighted
imaging
Jeff Kershaw1 and
Takayuki Obata1
1Applied MRI Research, QST, Chiba, Japan Keywords: Signal Representations, Diffusion/other diffusion imaging techniques, OGSE The target of oscillating-gradient spin-echo diffusion-weighted imaging (OGSE-DWI) is the spectral density of molecular diffusion, $$$u_2(\omega)$$$, which is predicted to obey a set of asymptotic universality relations that are linked to the global organisation of the sample. In principle, the complex microstructure of a medium can be classified by measuring the spectral density in its low- and high-frequency limits. However, the quantity most often measured with OGSE-DWI is the apparent diffusion coefficient (ADC). It was the purpose of this study to investigate how the low-frequency asymptotic behaviour of the spectral density is reflected in the ADC. |
||
5275.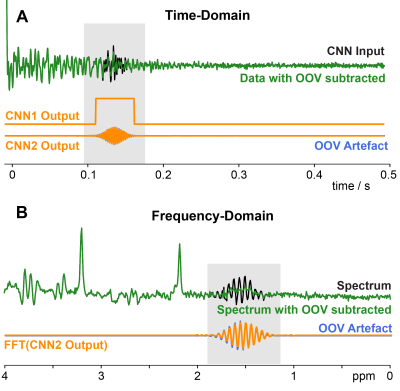 |
Using Convolutional Neural Networks to detect and remove
out-of-voxel MRS artefacts
Aaron T Gudmundson1,2,
Kathleen E Hupfeld2,3,
Yulu Song2,4,
Helge J Zöllner1,2,
Christopher W. Davies- Jenkins1,2,
İpek Özdemir1,2,
Georg Oeltzschner2,3,
and Richard A E Edden1,2
1The Russell H. Morgan Department of Radiology and Radiological Sciences, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3The Russell H. Morgan Department of Radiology and Radiological SciencesRadiology and Radiological Sciences, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 4The Russell H. Morgan Department of Radiology and Radiological Sciences Radiology and Radiological Sciences, Johns Hopkins University School of Medicine, Baltimore, MD, United States Keywords: Machine Learning/Artificial Intelligence, Spectroscopy, Out-of-voxel (OOV), MRS, Artefacts, Deep Learning, Convolutional Neural Network Out-of-voxel (OOV) artefacts, or echoes, are common in-vivo artefacts seen in MRS data. These artefacts are typically not identified until post-processing and are challenging to remove without modifying the underlying data. Here, we developed 2 Convolutional Neural Networks (CNNs) to overcome OOV artefacts at different stages. The first network (CNN1) was designed to identify OOV artefacts in single average data and offer a real-time assessment during data acquisition. The second (CNN2) predicts the OOV artefact to subtract during post-processing without impacting the metabolite data. |
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.