-
Multi-Scale Low-Rank Reconstruction for Phase-Cycled Projection-Reconstruction bSSFP Cardiac Cine and BMART-Generated B0 Maps
Anjali Datta1, Dwight Nishimura1, and Corey Baron2
1Electrical Engineering, Stanford, Stanford, CA, United States, 2Medical Biophysics, Western University, London, ON, Canada
Multi-scale low-rank reconstruction recovers high-quality phase-cycle images from a highly-undersampled frequency-modulated bSSFP cardiac cine sequence. It also facilitates estimation of a time series of B0 maps from the same data, which are used to combine the phase-cycles into the cine.
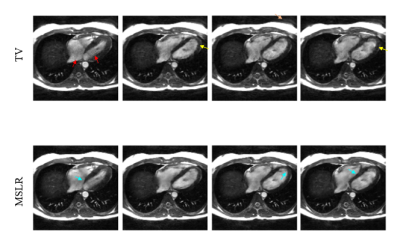
Field-map-combined
images for selected cardiac phases. The TV reconstructions have near-band-flow-related
streaking artifacts (red arrows) and undersampling artifacts (peach
arrow). In addition, some hyperintensities (yellow arrows) persist, possibly from using noisier field maps for
phase-cycle combination. The MSLR reconstructions have smoother blood signal since flow artifacts were localized to near
the bands, which were then removed by field-map combination. In addition, both blood-myocardium boundaries
(cyan arrows) and background structures appear sharper.
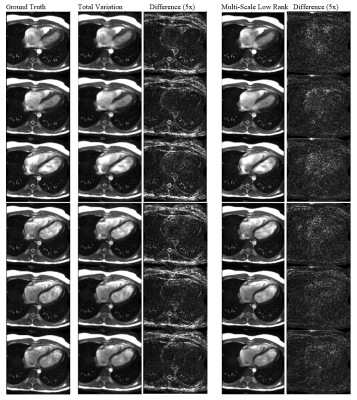
Reconstructions
from the inverse-gridded and retrospectively undersampled dataset. Six cardiac phases evenly spaced through the cardiac
cycle are shown. The phase cycles were
root-sum-of-squares combined. Multi-scale
low rank results in sharper images and better removal of undersampling
artifacts (e.g., in the background) than total variation.
-
Simplified Phase-Sensitive Inversion Recovery (PSIR) Reconstruction using Multi-dimensional Integration (MDI) for Elevated SNR
Yichen Hu1 and Junpu Hu2
1UIH America, Inc., Houston, TX, United States, 2United Imaging Healthcare, Shanghai, China
We applied
MDI algorithm to classic PSIR reconstruction for cardiac imaging and demonstrated
the effectiveness of the approach for improved SNR. In comparison to the conventional reconstruction, the algorithm
offers a simplified and fast pathway to achieve desired image contrast.
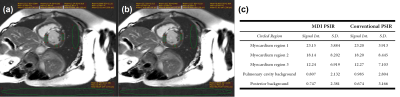
Figure
2. Images for detecting myocardial infarction by (a) MDI
PSIR and (b) the conventional PSIR reconstruction methods. (c) signal intensity
analysis for the five representative regions (three in myocardium and two in
the background) circled in (a) and correspondingly in (b).
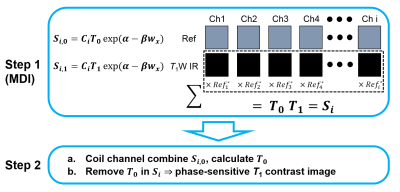
Figure
1. Illustration of MDI PSIR
reconstruction method
-
Single ProjectIon DrivEn Real-time (SPIDER) Multi-contrast MR Imaging Using Pre-learned Spatial Subspace
Pei Han1,2, Junzhou Chen1,2, Fei Han3, Zhehao Hu1,2, Debiao Li1,2, Anthony G. Christodoulou1,2, and Zhaoyang Fan1,4
1Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Department of Bioengineering, UCLA, Los Angeles, CA, United States, 3Siemens Medical Solutions USA, Inc., Los Angeles, CA, United States, 4Departments of Radiology and Radiation Oncology, University of Southern California, Los Angeles, CA, United States
A new technique called SPIDER is proposed for real-time multi-contrast 3D imaging. With the information learned and stored in the “Prep” scan, 3D multi-contrast images can be generated in the "Live" scan with simple matrix multiplication, which yielding a latency of 50ms or less.
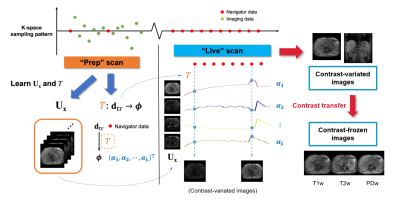
Figure 1: The SPIDER framework.
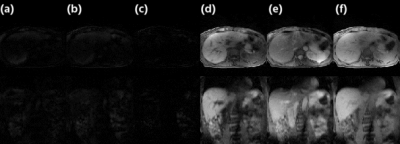
Figure 3: Comparison of images
from Multitasking reconstruction (reference) and the proposed SPIDER method.
From left to right: (a) Reference
from Multitasking reconstruction; (b) Contrast-variated SPIDER
real-time images; (c) Difference between (a) and (b); (d-f) Contrast-frozen
T1w, T2w, and PDw SPIDER real-time images.
-
Low-rank and Framelet Based Sparsity Decomposition for Reconstruction of Interventional MRI in Real Time
Zhao He1, Ya-Nan Zhu2, Suhao Qiu1, Xiaoqun Zhang2, and Yuan Feng1
1Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2School of Mathematical Sciences, MOE-LSC and Institute of Natural Sciences, Shanghai Jiao Tong University, Shanghai, China
A low-rank and sparsity decomposition with
framelet transform for spatial sparsity was proposed for reconstruction of
interventional MR images. A group-based reconstruction showed that the proposed
method can achieve an acceleration of 40 folds.
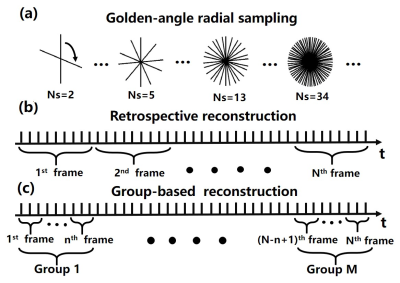
Figure 1.
Illustrations of the data acquisition and reconstruction scheme. (a) A
continuous golden-angle radial sampling method was used for i-MRI in this study
(golden angle = 111.25°). (b) Conventional dynamic image
reconstruction based on a retrospective scheme. (c) The proposed group-based
reconstruction method for real-time i-MRI reconstruction.
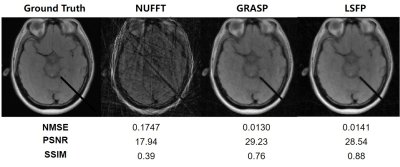
Figure 2. A
comparison of different algorithms. The ground truth is the 150th simulated
brain intervention image. A group-based reconstruction strategy (10 spokes per
frame, 5 frames per group, a total of 200 frames for 2000 spokes) was adopted
for reconstruction using NUFFT, GRASP, and LSFP. The acceleration factor was about
40.
-
Reconstruction of Undersampled Dynamic MRI Data Using Truncated Nuclear Norm Minimization and Sparsity Constraints
Runyu Yang1, Yuze Li1, and Huijun Chen1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China
Achieving high spatio-temporal resolutions is challenging in dynamic magnetic resonance imaging . It is effective to use low-rank prior and sparse prior for dMRI reconstruction. We proposed a novel method used low rank which utilize a nonconvex norm and sparse for dMRI reconstruction.
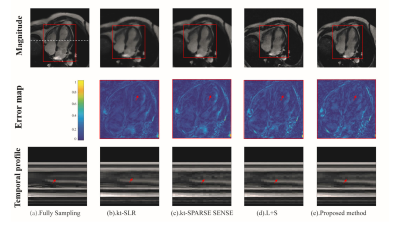
FIG.2. Reconstruction
results comparison for cardiac cine dataset with radial trajectory and r=2. From top to bottom: time-frame magnitude images, error map, x-t
temporal profiles in white dotted line. The proposed method had lower artifacts for
reconstruction of cardiac blood pools than the other methods by the arrows in the error map. By the arrows in the temporal profile, the
methods to be compared present temporal blurring artifacts, which are
effectively removed by the proposed mothed.
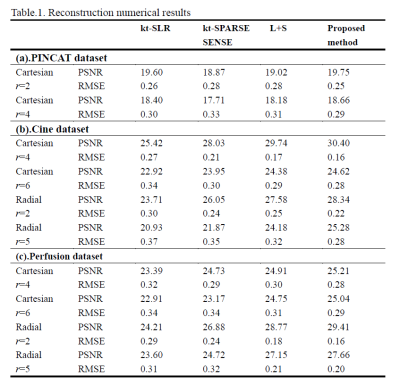
Table.1. PSNR/RMSE
comparison of various reconstruction methods on PINCAT dataset, Cine dataset
and Perfusion dataset.
-
Time Domain Principal Component Analysis for Rapid, Real-Time MRI Reconstruction from Undersampled Data
Mark Wright1, Bryson Dietz1, Jihyun Yun1,2, Eugene Yip2, B Gino Fallone1,2, and Keith Wachowicz1,2
1Oncology, University of Alberta, Edmonton, AB, Canada, 2Medical Physics, Cross Cancer Institute, Edmonton, AB, Canada
A real-time acceleration method using Principal Component Analysis (PCA) was developed for use on hybrid MR-radiotherapy machines. Good temporal-robustness was achieved at high frame-rate and low latency (less than 50ms), without the need for any complex coil geometries.
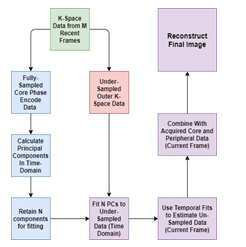
Figure 2: Flow chart of the
acceleration method. All calculations are done in the k-space domain.
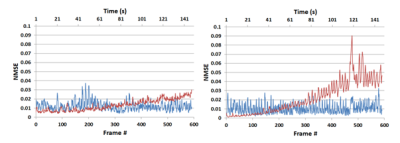
Figure 3: A time evolution
comparison of for two patients over 2.5 minutes using the spatial PCA method
(red) and our proposed time-domain PCA method (blue) using normalised mean
square error for an acceleration factor of 3.
-
Optimal Transport Based Convex Hybrid Image and Motion-Field Reconstruction
Ingmar Middelhoff1, Matthias Schlögl2, Adrián Martín Fernández3, Silvio Fanzon4,5, Kristian Bredies4,5, and Rudolf Stollberger1,5
1Institute of Medical Engineering, TU Graz, Graz, Austria, 2Solgenium OG, Linz, Austria, 3Department of Information and Communications Technologies, Pompeu Fabra University, Barcelona, Spain, 4Institute of Mathematics and Scientific Computing, NAWI Graz, University of Graz, Graz, Austria, 5BioTechMed-Graz, Graz, Austria
An Optimal Transport based reconstruction for motion-afflicted data is tested which yields an image series and pixel-wise motion fields. Reconstructions based on simulated single-coil 4-fold undersampled k-space time-series data show good image quality.
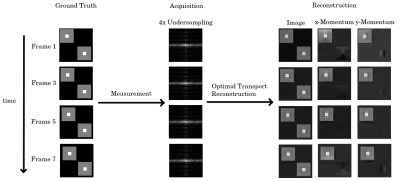
A visualization of the reconstruction concept: A moving object is measured in a series of undersampled k-spaces. In this abstract, we simulated a 4-times acceleration per k-space with the 8 center k-space lines being measured additionally. The OT reconstruction allows not only to recover the image-series, but also the momentum fields of the object. For illustration purposes, only every second frame is shown.
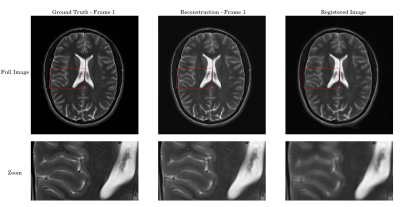
Zoom-in on the results from figure 2: The reconstructed image closely mirrors the ground truth. Despite every frame only having approximately one fourth of k-space sampled with a single coil, the brain could be reconstructed in detail with only the optimal transport regularization. The registered image is slightly blurry, showing potential problems with the motion field.
-
Accelerating gSlider-based Diffusion MRI: Phase constraints enable reduced RF encoding
Yunsong Liu1, Kawin Setsompop2, and Justin P. Haldar1
1Signal and Image Processing Institute, University of Southern California, Los Angeles, CA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States
We investigate whether smooth-phase constraints can be used
to reduce the required number of RF encodings in gSlider diffusion MRI. Theoretical and simulation results
demonstrate that, it can be done if optimized RF encodings are used.
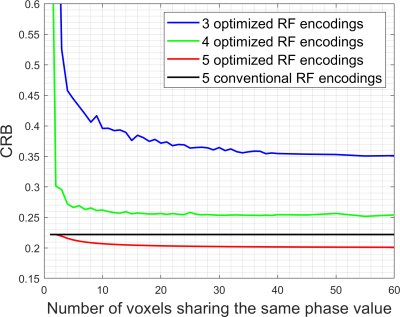
Figure 2. CRBs for resolving 5 thin
slices as a function of the number of voxels sharing the same phase value,
plotted for several different RF encoding strategies. Note that without phase constraints, the CRB
blows up to infinity whenever the number of RF encodings is smaller than the
number of thin slices (5 in this case).
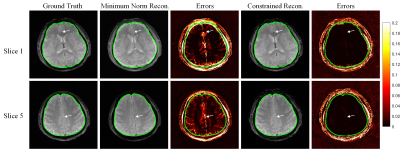
Figure 3. Simulated reconstruction of 5 thin slices from
the set of 4 optimized RF encodings, for both phase-constrained and minimum
norm reconstruction. For ease of visualization, we only show two out of the five
reconstructed slices for each case.
-
Improved Sampling for Distortionless Diffusion Weighted 2D Cartesian Multi-Shot Fast Spin Echo
Philip Kenneth Lee1,2, Yuxin Hu1,2, Catherine Judith Moran2, Bruce Lewis Daniel2, and Brian Andrew Hargreaves1,2,3
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States, 3Biomedical Engineering, Stanford University, Stanford, CA, United States
Cartesian Fast Spin
Echo can improve distortionless diffusion weighted imaging with multi-shot sampling
patterns that complement low-rank compressed sensing reconstructions.
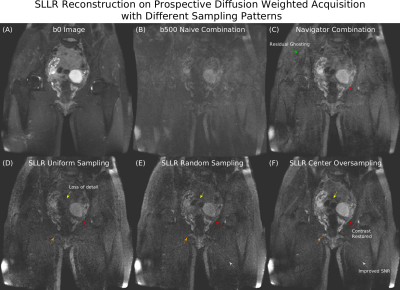
Figure 5: (A) T2-weighted b0 image. (B) Naive combination of 12-shot data has severe multi-shot
ghosting resulting in signal dropout. (C) Navigator shot combination has some residual ghosting (green). (D)
SLLR reconstruction of uniformly sampled b500 data has loss of detail (yellow,
orange), and reduced organ contrast (red). (E) Random sampling improves detail
(yellow, orange). (F) Center oversampling further enhances detail (yellow,
orange), restores organ contrast, matching the navigator combination (red), and
improves SNR (white).
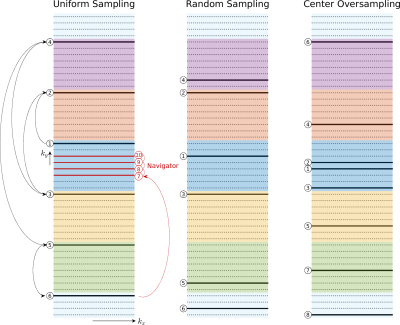
Figure
1: Random sampling patterns were generated by dividing
k-space into $$$ETL$$$ segments, represented by different colours. Each sample is
randomly assigned to a shot. K-space is fully covered when shots are combined.
For center oversampling, the center two ky lines were added to the beginning of
a randomly generated shot. A low-resolution phase navigator was explicitly acquired
with uniformly sampled data because the shot undersampling factor is too high
for parallel imaging to recover phase maps.
-
Model-Based Iterative Reconstruction for Short-Axis Propeller EPI at 7T MRI
Uten Yarach1,2, Frank Godenschweger3, Matt A Bernstein2, Myung-Ho In2, Itthi Chatnuntawech44, Kawin Setsompop5, Oliver Speck3, and Joshua Trzasko2
1Radiologic Technology Department, Associated Medical Sciences, Chiang Mai University, Chinag Mai, Thailand, 2Department of Radiology, Mayo Clinic, Rochester, MN, USA, Rochester, MN, United States, 3Otto-von-Guericke University Magdeburg, Biomedical Magnetic Resonance, Magdeburg, Germany, 4National Nanotechnology Center (NANOTEC), National Science and Technology Development Agency (NSTDA), Bangkok, Thailand, 5Department of Radiology, Stanford University, Stanford, CA, United States
Model-based iterative reconstruction can manage for
off-resonance effect. This framework with locally-low-rank regularization enables high-resolution SAP-EPI images with minimizing blurring artifact.
Moreover, no phase-calibration of different multi-blade directions is required.
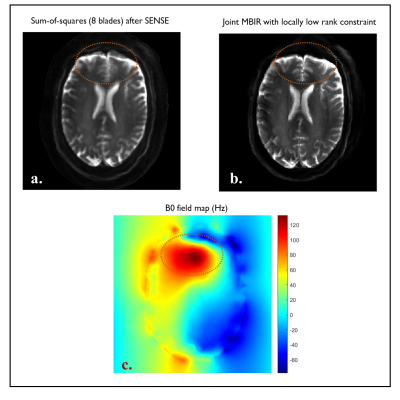
Fig. 3. (a): Sum-of-squares image obtained from all individual
blade images in 2a. (b): The image obtained by the proposed MBIR with virtual
coil LLR. (c): B0 field map obtained from 6-echo fast gradient-echo images via
fat-water separation. The red circles highlight severe geometric distortion (a)
and its improvement with the proposed scheme (b) in the region with strong
field inhomogeneity as shown in (c).
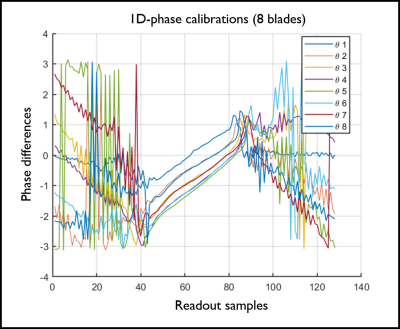
Fig.1.
1D
phase differences between positive and negative readout echoes (2x-oversampling
not removed). Their offsets and slopes appear slightly
different among the eight different blade angles.
-
On the possibility of reconstructing arbitrary FOVs using gradient waveforms with low-coherent aliasing properties
Tobias Speidel1, Patrick Metze1, Kilian Stumpf1, Thomas Hüfken1, and Volker Rasche1
1Internal Medicine II, Ulm University Hospital, Ulm, Germany
The calculation of k-space trajectories in MRI usually involves prior knowledge of the FOV due to a minimum k-space sampling density. Based on a generalised form of the "Seiffert Spirals", this abstract describes an imaging modality that does not require prior commitment to an imaging FOV.
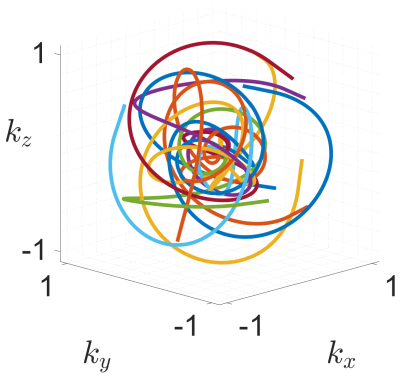
Figure 1: 15 interleaves of the generated trajectory within normalised k-space.
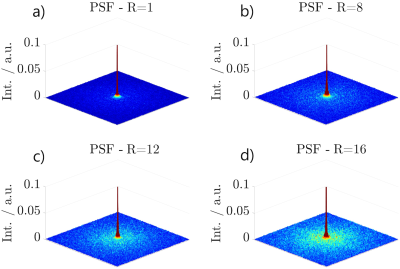
Figure 2: Simulated PSFs in the $$$xy$$$-plane with $$$z=0$$$ of the presented approach according to the undersampling factors $$$R_1$$$ in a), $$$R_2$$$ in b), $$$R_3$$$ in c) and $$$R_4$$$ in d).
-
Learning a Preconditioner to Accelerate Compressed Sensing Reconstructions
Kirsten Koolstra1 and Rob Remis2
1Division of Image Processing, Leiden University Medical Center, Leiden, Netherlands, 2Circuits and Systems, Delft University of Technology, Delft, Netherlands
In this work we design a preconditioner for compressed sensing reconstructions using a neural network. Results show that it is possible for a learned preconditioner to improve upon the performance of existing preconditioning techniques.
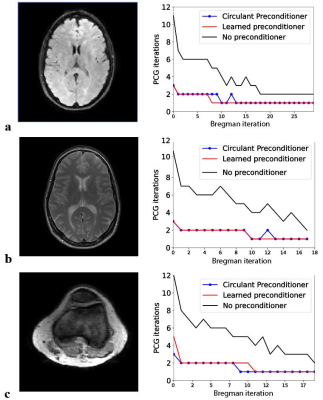
Fig.2. Comparison of CG convergence with and without the learned and the circulant preconditioner. (a) Flair brain scan (128x128). The learned preconditioner reduces the number of iterations in CG by a factor 2.9. This is a slightly larger reduction compared to the factor of 2.7 obtained with the circulant preconditioner. (b) Similar results are obtained for a TSE scan with a twice as large matrix size compared to that in the training set. (c) The speed up is slightly lower in the knee (128x128), which is an anatomy that the network has not observed during training.
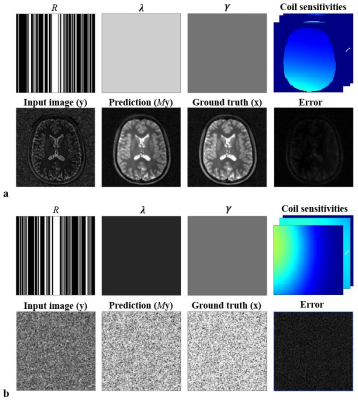
Fig.1. The network’s input and output for two training examples. Each 37-channel input contains a sampling mask (R), regularization masks (λ and γ), 16 complex coil sensitivity maps and a complex image (y). Note that the complex channels are first split into real and imaginary components. The network’s prediction My is close to the ground truth, which is confirmed by the small error values both for the brain case (a) and for the noise case (b) (normalized norm < 0.16).
-
Robust and Computationally Efficient Missing Point and Phase Estimation for Zero Echo Time (ZTE) Sequences
Curtis A Corum1,2, Abdul Haseeb Ahmed2, Mathews Jacob2, Vincent Magnotta2, and Stanley Kruger2
1Champaign Imaging LLC, Shoreview, MN, United States, 2University of Iowa, Iowa City, IA, United States
Here
we modify and apply for the first time a robust and computationally efficient missing point and
phase estimation algorithm originating in the solid state NMR
community for zero echo time (ZTE) imaging sequences.
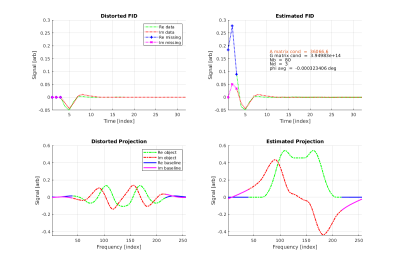
Figure
1: Distorted FID, distorted
Projection, corrected FID
and corrected projection fro simulated hollow cube object.
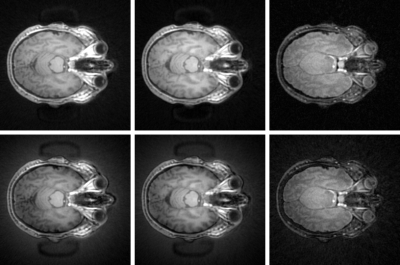
Figure
2: Brain Images of Health Adult
Subject.
Bottom Row Left to
Right: Un-corrected ZTE intrinsic contrast, IR-ZTE and T2-ZTE
Top
Row Left to Right: Corrected
(Nd = 3) ZTE
intrinsic
contrast, IR-ZTE and T2-ZTE
-
Iterative Reconstruction for Enhanced Through-Plane Resolution T2-Weighted Spin-Echo Imaging of the Prostate
Eric A Borisch1, Roger C Grimm1, Soudabeh Kargar2, Akira Kawashima3, Joshua D Trzasko1, and Stephen J Riederer1
1Radiology, Mayo Clinic, Rochester, MN, United States, 2Radiology, University of Wisconsin-Madison, Madison, WI, United States, 3Radiology, Mayo Clinic, Phoenix, AZ, United States
A sparsity-regularized forward model-based iterative reconstruction is presented improving the through-plane resolution and noise performance of the output images produced from a set of overlapping lower (thicker) resolution 2D acquisitions.
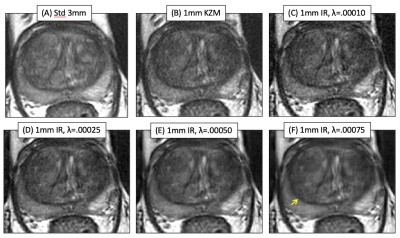
Axial T2SE images of the prostate of one subject: (A) standard 3mm thick T2SE (acquired in 4:14) and (B-F) 1mm thick images. (B) was reconstructed using the linear KZM method[1], while (C-F) were generated with the new MBIR at the λ values shown. Note some improved sharpness of (B) vs. (A) owing to the thinner slice but an increased noise level. MBIR with increasing λ provides progressive reduction in noise in (C-F). However, use of λ=0.00075 caused undesirable blurring, e.g. between the peripheral and transition zones (F, arrow).
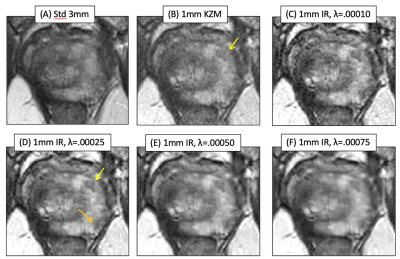
Results from a second case. Fig. 2B vs. A better portrays a lesion (B, arrow), but MBIR provides further improved depiction (D, yellow arrow) and definition of a second lesion (orange arrow). Higher λ values again cause blur. Across the patient studies the radiologist’s perceived preference was for λ=0.00025.
-
Automatic WaveCS reconstruction
Gabriel Varela-Mattatall1,2 and Ravi S Menon1,2
1Centre for Functional and Metabolic Mapping, Robarts Research Institute, Western University, London, ON, Canada, 2Department of Medical Biophysics, Schulich School of Medicine and Dentistry, Western University, London, ON, Canada
We present an automatic, non-iterative, and prospective method to determine the regularization weighting for WaveCS reconstructions. The image quality from this reconstruction is on the same order as other reconstruction but without any tuning.
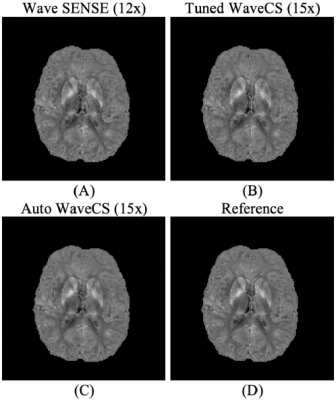
Visualization of local phase images from the different reconstruction procedures. Panels (A) and (B) show the local phase from Bilgic et al7, while panel (C) shows the local phase obtained from our automatic WaveCS reconstruction procedure. Panel (D) shows the reference. In parenthesis it is shown the under-sampling factor for each reconstruction.
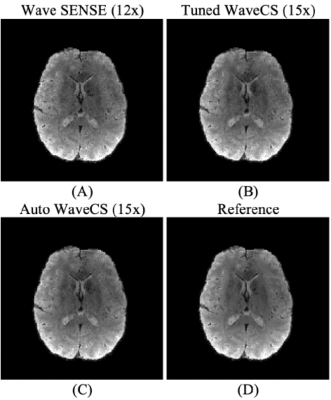
Visualization of magnitude Images from the different reconstruction procedures. Panels (A) and (B) show the results from Bilgic et al7, while panel (C) shows the result from our automatic WaveCS reconstruction. Panel (D) shows the reference. In parenthesis it is shown the under-sampling factor for each reconstruction.
-
Jointly Reconstructed Undersampled Multiparameter MRI for Imaging Intratumoral Subpopulations
Shraddha Pandey1,2, Arthur David Snider1, Wilfrido Moreno1, Harshan Ravi2, Ali Bilgin3, and Natarajan Raghunand2,4
1Electrical Engineering, University of South Florida, Tampa, FL, United States, 2Cancer Physiology, Moffitt Cancer Center, Tampa, FL, United States, 3Departments of Medical Imaging, Biomedical Engineering, and Electrical & Computer Engineering, University of Arizona, Tucson, AZ, United States, 4Department of Oncologic Sciences, University of South Florida, Tampa, FL, United States
A joint reconstruction framework is presented to concurrently reconstruct a series of complex MR images and their corresponding T1, T2 and T2* parameter maps. Tissue mapping and estimation of water and fat content within 4 objectively defined tissue types was possible using 18% k-space data.
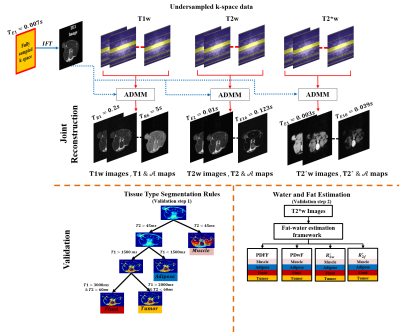
Figure 1. k-space data
obtained from the scanner is undersampled using the cartesian sampling mask.
The Joint Reconstruction Algorithm is used to reconstruct the series of T1w
images and their parameter maps. The process is repeated to reconstruct
T2w/T2*w images and their parameter maps. Validation of the results is carried
out by identifying the tissue types like muscle, fluid, tumor and adipose and using the rules on T1 and T2 maps. The T2*w images are subjected [1]
to estimate PDFF, PDwF, & in the muscle, fluid, tumor and adipose tissue
type.
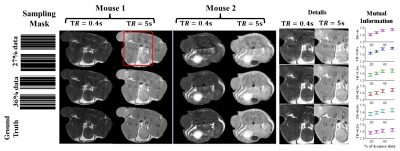
Figure 2. T1w
reconstructed images for 2 mouse slices with mask of ~27%, ~36% k-space
data are shown.The sampling masks are shown in col 1. The results
for the Repetition times (TR) 0.4s and 5s are shown in col 2 & 3 for mouse 1 and col 4 & 5 for mouse 2. The 5th
& 6th column shows the detailed version of the region
highlighted in the red box.The MI value is computed for 4 undersampling masks 18%, 27%, 36%, & 52% shown on x-axis. The y-axis shows the MI value when the ground truth |u| are compared to the reconstructed |u|. The mean MI and standard error of the mean (S.E.M) is calculated over n = 30 mouse slices.
-
Automatic determination of the regularization weighting for wavelet-based compressed sensing MRI reconstructions
Gabriel Varela-Mattatall1,2, Corey A Baron1,2, and Ravi S Menon1,2
1Centre for Functional and Metabolic Mapping, Robarts Research Institute, Western University, London, ON, Canada, 2Department of Medical Biophysics, Schulich School of Medicine and Dentistry, Western University, London, ON, Canada
Here we present an automatic, non-iterative, prospective, and fast approach to determine the regularization weighting, which enables wavelet-based compressed sensing MRI reconstructions.
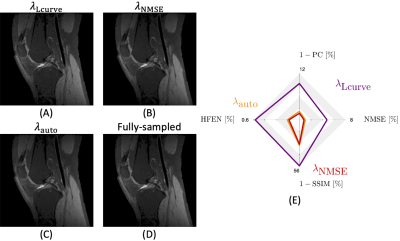
Knee images from the different strategies to define the regularization weighting. We compare (A) $$$ \lambda_{\textrm{Lcurve}} $$$ (14 iterations) and (B) $$$ \lambda_{\textrm{NMSE}} $$$ (14 iterations) strategies to (C) our approach, $$$ \lambda_{\textrm{auto}} $$$, using the 4th-level Daubechies mother wavelet (1 iteration). Panel (D) shows the reference and panel (E) shows quantitative results. Smaller values along each axis in (E) represent more accurate reconstructions.
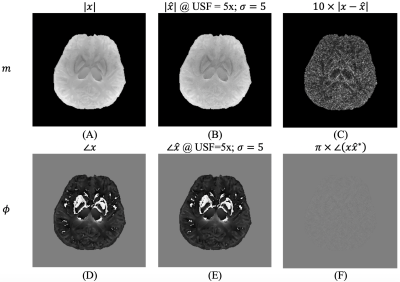
Visualization of magnitude and phase images from simulations. Panels (B) and (E) show the images from the reconstruction process using our approach for USF=5 and $$$\sigma=5$$$. Panels (A) and (D) show the reference. Panels (C) and (F) show the difference between both columns. All images are scaled according to their respective reference.
-
Improved CS-MRI using Hybrid Plug-and-Play Priors based Fast Composite Splitting Algorithm
Qingyong Zhu1, Jing Cheng2, Zhuo-Xu Cui1, and Dong Liang1,2
1Research Center for Medical AI, SIAT, Chinese Academy of Sciences, Shenzhen, China, 2Paul C. Lauterbur Research Center for Biomedical Imaging, SIAT, Chinese Academy of Sciences, Shenzhen, China
The proposed hybrid Plug-and-Play priors (PnP) based on internal and external denoising can help improve the performance of fast composite splitting algorithm in MRI, and the hybrid PnP is also easily incorporated in other algorithms such as ADMM and AMP.
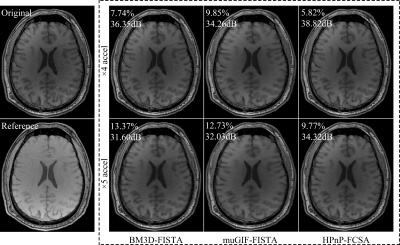
Figure 1. The reconstruction results using all comparison methods at AF= 4, 5. The values at left-top of images are the corresponding RE($$$\%$$$) and PSNR(dB).
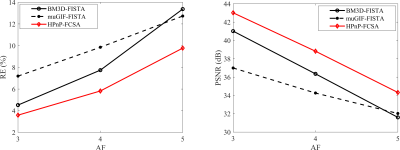
Figure 2. The RE (left) and PSNR (right) plots of all reconstructions at various AFs (AF=3, 4, 5).
-
Fast Variable Density Poisson-Disc Sample Generation with Directional Variation for Compressed Sensing
Nicholas Dwork1, Corey A. Baron2, Ethan M. I. Johnson3, Daniel O'Connor4, John M. Pauly5, and Peder E.Z. Larson1
1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Robarts Research, Western University, Ontario, ON, Canada, 3Biomedical Engineering, Northwestern University, Evanston, IL, United States, 4Mathematics and Statistics, University of San Francisco, San Francisco, CA, United States, 5Electrical Engineering, Stanford University, Stanford, CA, United States
We have developed a fast algorithm to create a variable density poisson disc sampling pattern.
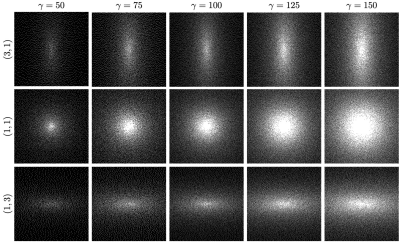
Fig. 2: Variable density poisson disc sampling patterns for different values of $$$\gamma$$$ and different aspect ratios.
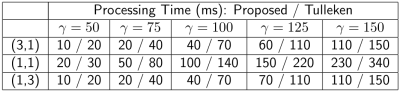
Table 1: Run times for generating variable density poisson-disc sampling patterns with the proposed algorithm and the Tulleken algorithm. Time is reported in milliseconds. In all cases, the proposed algorithm is faster by $$$30-50\%$$$.
-
Automatic determination of the regularization weighting for low rank reconstruction problems
Gabriel Varela-Mattatall1,2, Corey A Baron1,2, and Ravi S Menon1,2
1Centre for Functional and Metabolic Mapping, Robarts Research Institute, Western University, London, ON, Canada, 2Department of Medical Biophysics, Schulich School of Medicine and Dentistry, Western University, London, ON, Canada
We develop a general, non-iterative, fast, and automatic procedure to determine the regularization weighting for low-rank reconstruction problems.
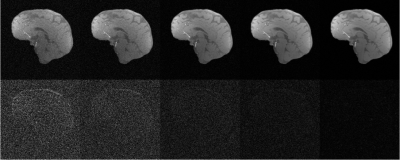
Impact of the size of the dataset for automatic low rank reconstructions using SNR 5 and an under-sampling factor of 4x. The first row corresponds to the mean appearance of the reconstructions, $$$\hat{X}$$$, for 5,10,40,100, and 600 images (from left to right, respectively). The second row corresponds to the absolute difference with respect to the reference, $$$X$$$, as $$$5 \times |\hat{X}-X|$$$.
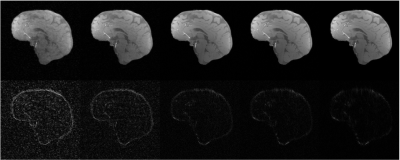
Impact of the size of the dataset for automatic low rank reconstructions using SNR 30 and an under-sampling factor of 14x. The first row corresponds to the mean appearance of the reconstructions, $$$\hat{X}$$$, for 5,10,40,100, and 600 images (from left to right, respectively). The second row corresponds to the absolute difference with respect to the reference, $$$X$$$, as $$$5 \times |\hat{X}-X|$$$.
