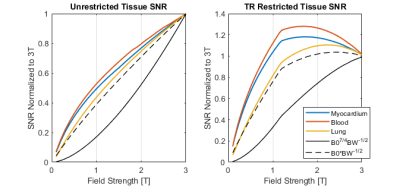
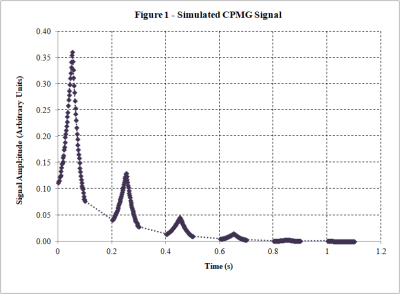
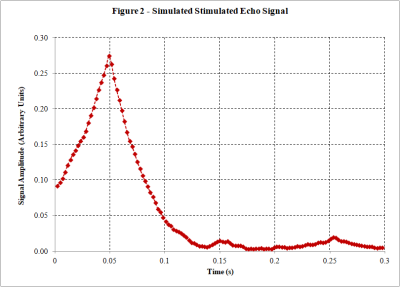
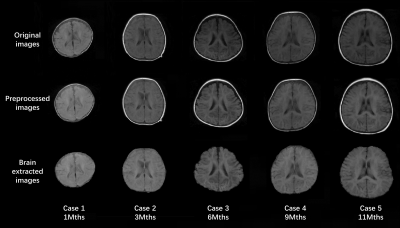
-
qMTNet+: artificial neural network with residual connection for accelerated quantitative magnetization transfer imaging
Huan Minh Luu1, Dong-Hyun Kim1, Seung-Hong Choi2, and Sung-Hong Park1
1Magnetic Resonance Imaging Laboratory, Department of Bio and Brain Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of
In this study, we propose qMTNet+, an improved version of qMTNet that accomplishes acceleration for data acquisition and fitting , as well as generation of missing data with a single residual network. Results showed that qMTNet+ improves the quality of generated MT images and qMT parameters.
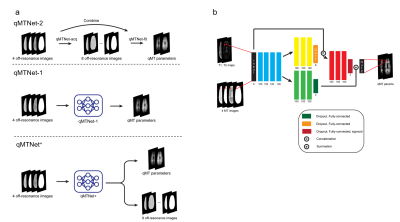
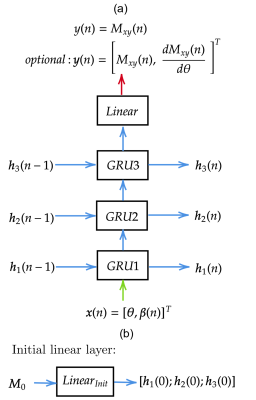
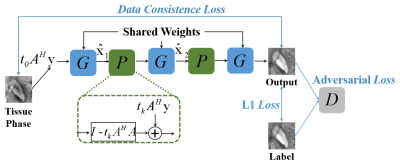
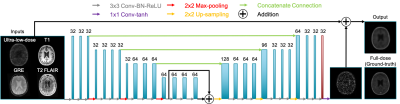
Figure 1: a) Overview of qMTNet-2,
qMTNet-1 and qMTNet+
approach:
qMTNet-2 comprises of 2 separate sub-networks. qMTNet-1 is a single
integrated network to directly predict qMT parameters from undersampled MT
images. qMTNet+ consists of a single network that can
produce both values of interest. b) Structure of qMTNet+. Unless stated in the caption, the layer includes 100 hidden neurons
with rectified linear unit (ReLU) activation and batch normalization.
Different color signifies different computation path in the network.
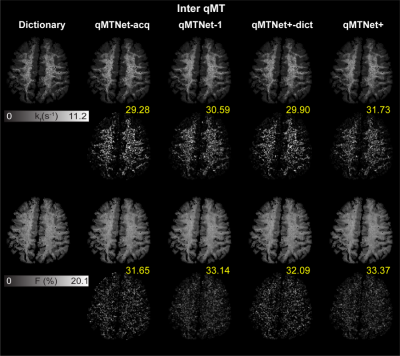
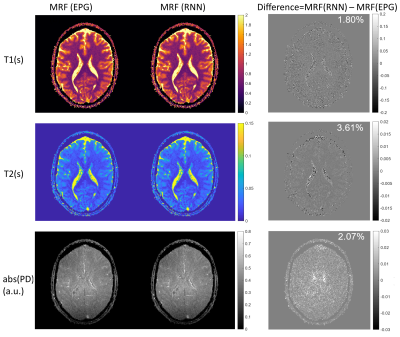
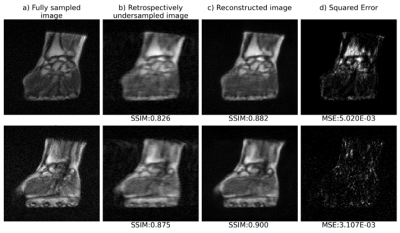
Figure 3: Qualitative comparison
between qMTNet+ and qMTNet
output against dictionary fitted qMT parameters on inter-slice MT data.
Dictionary
specifies qMT
parameters obtained from dictionary fitting of acquired MT data and is
considered as the label. The numbers
at the bottom of the images are the peak signal-to-noise ratio. The top two
rows show result for kf and the bottom two rows show result for
F. The first row shows qMT
parameters from the different fitting methods and the second row shows the 5
times magnified absolute differences. Details of the networks are explained in text.
-
Global and Local Deep Dictionary Learning Network for Accelerating the Quantification of Myelin Water Content
Quan Chen1, Huajun She1, Zhijun Wang1, and Yiping P. Du1
1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China
A Global and Local
Deep Dictionary Learning Network (GLDDL) is proposed to accelerate the MWF
mapping. The GLDDL network explores both global and local spatiotemporal
correlations. The merits of traditional DL and that of the deep learning are
combined in the propose network.
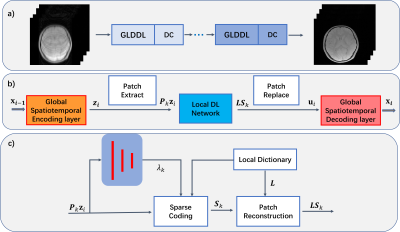
Figure
1. a) The overview of the propose GLDDL network. b) The GLDDL block. The global
spatiotemporal encoder and decoder layers are used before and after the local
DL network, respectively. c) The details
of the local DL network.
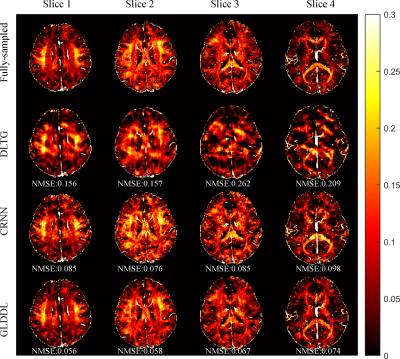
Figure
3. The comparative MWF maps of the DLTG, CRNN, and GLDDL reconstructions at R =
6 from one subject. The NMSE are showed at the bottom of the corresponding images.
-
Rapid MR Parametric Mapping using Deep Learning
Jing Cheng1, Yuanyuan Liu1, Xin Liu1, Hairong Zheng1, Yanjie Zhu1, and Dong Liang1
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
In this
work, we proposed to incorporate the quantitative
physical model into the deep learning framework to simultaneously reconstruct
the parameter-weighted images and generate the parametric map without the
reference parametric maps.
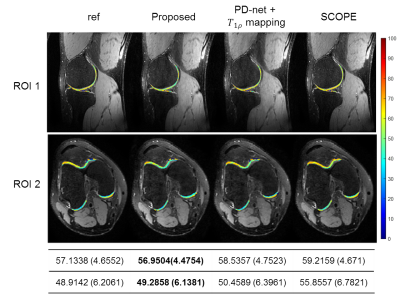
Figure
4. The estimated parametric maps for selected cartilage ROIs overlaid
on the reconstructed T1ρ-weighted
images at TSL = 5 ms for R = 9.2 from Data 2. The mean values and the standard deviations
of the ROI maps are also provided.
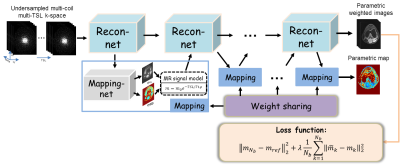
Figure
1. Overview
of the proposed framework.
-
Synthesizing large scale datasets for training deep neural networks in quantitative mapping of myelin water fraction
Serge Vasylechko1,2, Simon K. Warfield1,2, Sila Kurugol1,2, and Onur Afacan1,2
1Boston Children's Hospital, Boston, MA, United States, 2Harvard Medical School, Boston, MA, United States
We generated substantial amount of 3D synthetic T2 relaxometry data with a realistic forward model, and demonstrate its application to myelin water fraction. Our network has resulted in an excellent accuracy in the synthetic test dataset, and generated similar MWF maps as the NNLS algorithm.
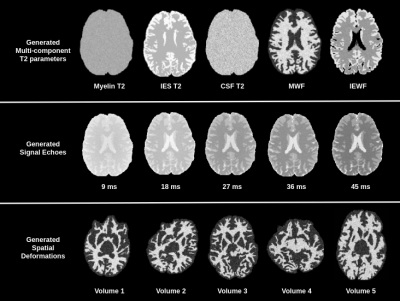
Figure 2: An example of the generated synthetic data. Top row shows the model parameters, second row shows the generated signals and third row shows generated spatial transformations.
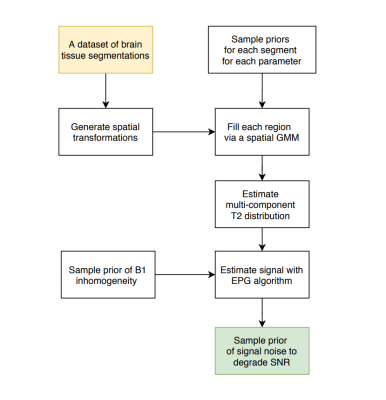
Figure 1: A flowchart detailing the proposed pipeline for generation of large scale 3D synthetic datasets for multi-component T2 distributions within the naturally occuring bounds, with a spatially varying sampling model.
-
Deep unrolled network with optimal sampling pattern to accelerate multi-echo GRE acquisition for quantitative susceptibility mapping
Jinwei Zhang1, Hang Zhang1, Pascal Spincemaille2, Mert Sabuncu3, Thanh Nguyen2, Ilhami Kovanlikaya2, and Yi Wang2
1Cornell University, New York, NY, United States, 2Weill Cornell Medical College, New York, NY, United States, 3Cornell University, Ithaca, NY, United States
An unrolled ADMM reconstruction network with learned optimal sampling pattern is trained to accelerate multi-echo 3D-GRE acquisition for quantitative susceptibility mapping. Prospective study shows the learned pattern achieves better QSM image quality than a manually designed pattern.
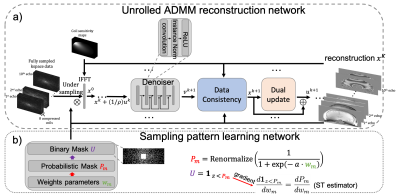
Figure 1. Proposed network architecture. Unrolled ADMM network (a) reconstructed multi-echo images from the under-sampled multi-coil multi-echo kspace data. Sampling pattern learning network (b) optimized the kspace undersampling pattern with a straight-through estimator to improve back-propagation.
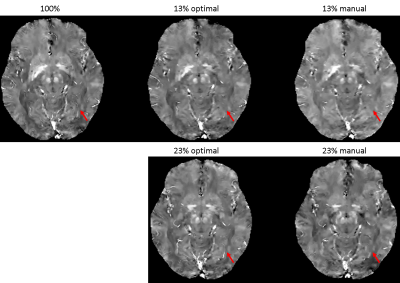
Figure 3. QSMs of optimal and manually designed sampling patterns with 13% and 23% sampling ratios, with fully sampled (100%) QSM as the reference. QSM sampled by the optimal pattern captures more details in the image such as veins (red arrow) compared to the manual pattern. With 13% under-sampling ratio, both QSMs by the optimal and manual patterns lose some detailed structures (red arrow).
-
Automated quantitative evaluation of deep learning model for reduced gadolinium dose in contrast-enhanced brain MRI
Srivathsa Pasumarthi1, Jon Tamir2, Enhao Gong2, Greg Zaharchuk2, and Tao Zhang2
1Subtle Medical Inc, Menlo Park, CA, United States, 2Subtle Medical Inc., Menlo Park, CA, United States
This work proposes an automated quantitative evaluation scheme for the GBCA dose reduction using DL.

Overall processing pipeline for the quantitative evaluation scheme. The post-contrast 3D T1W (CE) and DL-CE volumes were skull-stripped, interpolated and co-registered to anatomical template. The volumes are then processed with the BRATS pre-trained volume through the NGC interface to obtain the Tumor Core (TC) segmentation masks.
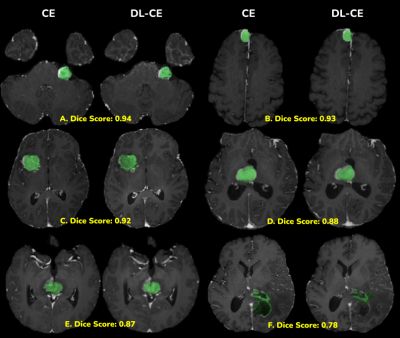
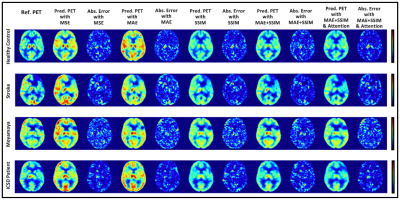
CE and DL-CE shown side-by-side with the segmented Tumor Core (TC) (green overlay). Individual Dice scores are shown below each image pair. The segmentation mask of DL-CE in image (F) is different from that of CE, even though the enhancement patterns look similar.
-
Using an ANN to estimate Initial Values for Mapping of the Oxygen Extraction Fraction with combined QSM and qBOLD
Patrick Kinz1, Sebastian Thomas1, and Lothar R. Schad1
1Computer Assisted Clinical Medicine, Heidelberg University, Medical Faculty Mannheim, Mannheim, Germany
Combining an ANN with traditional quasi-Newton methods increases robustness an reduces noise of OEF mapping with QSM and qBOLD.
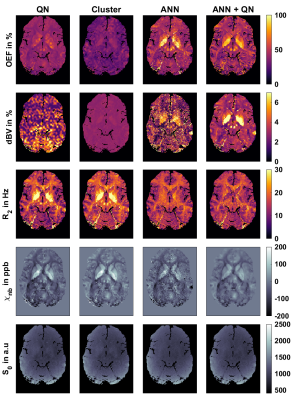
Fig.1: Representative axial slice of the reconstructed parameters: oxygen extraction fraction OEF, deoxygenated blood volume dBV, transverse relaxation rate R2, non-blood susceptibility χnb and magnitude after excitation S0. Reconstruction is based on combined QSM and qBOLD and done with four different approaches: a voxel wise quasi-Newton (QN) approach, QN initialized with the results from an initial fit on clusters of voxels with similar signal development (Cluster), an artificial neural network (ANN), and QN initialized with the results from the ANN (ANN+QN).
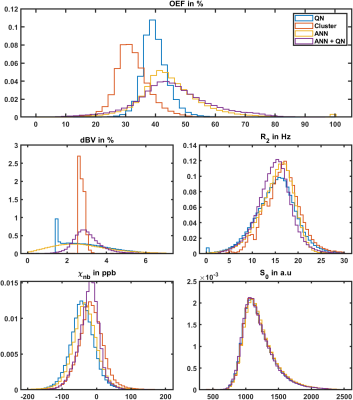
Fig.2: Normalized histograms of reconstructed parameters in gray matter (left) and white matter (right) of the same test subject as in figure 1. Parameters are oxygen extraction fraction OEF, deoxygenated blood volume dBV, transverse relaxation rate R2, non-blood susceptibility χnb and magnitude after excitation S0. Reconstruction approaches are: voxel wise quasi-Newton (QN), QN initialized with an initial fit on clusters of voxels with similar signal development (Cluster), an artificial neural network (ANN), and QN initialized with the results from the ANN (ANN+QN).
-
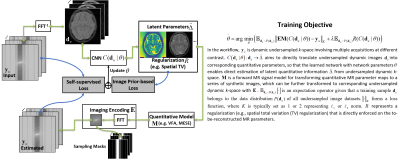
A self-supervised deep learning approach to synthesize weighted images and T1, T2, and PD parametric maps based on MR physics priors
Elisa Moya-Sáez1,2, Rodrigo de Luis-García1, and Carlos Alberola-López1
1University of Valladolid, Valladolid, Spain, 2Fundación Científica AECC, Valladolid, Spain
- - A self-supervised deep learning approach to compute T1, T2, and PD maps from clinical routine sequences.
- - Any realistic weighted images can be synthesized from the parametric maps.
- - The proposed self-supervised CNN achieves significant improvements in most synthesized modalities.
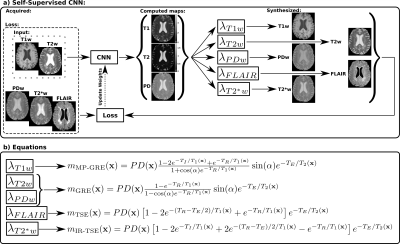
Figure 1: Overview of the proposed approach. a) Pipeline for training and testing the self-supervised convolutional-neural-network (CNN). b) Equations of the lambda layers used to synthesize the T1w, T2w, PDw, T2*w and FLAIR images from the previously computed parametric maps and with sequence parameters of Table 1. m(x) is the signal intensity of the corresponding weighted image at pixel x. Note that the CNN is pre-trained exclusively with synthetic data, as described in Ref7, and that PDw, T2*w, and FLAIR images are only used to compute the loss function and are not input to the CNN.
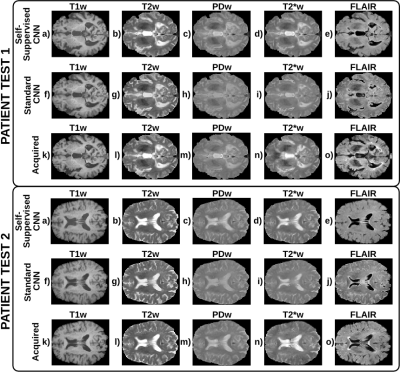
Figure 3: A representative axial slice of the synthesized and the corresponding acquired weighted images for each test patient. a-e) T1w, T2w, PDw, T2*w, and FLAIR images synthesized by the self-supervised CNN. f-j) Corresponding images synthesized by the standard CNN. k-o) Corresponding acquired images. The self-supervised CNN achieves better structural information and contrast than the standard CNN obtaining higher quality synthesis. Specifically, for the PDw, T2*w, and FLAIR which had the lowest quality synthesis in the standard CNN. See the CSF of the PDw, T2*w, and FLAIR.
-
Free-breathing Abdomen T2 mapping via Single-shot Multiple Overlapping-echo Acquisition and Deep Neural Network Reconstruction
Xi Lin1, Qinqin Yang1, Jianfeng Bao2, Shuhui Cai1, Zhong Chen1, Congbo Cai1, and Jingliang Cheng2
1Department of Electronic Science, Xiamen University, Xiamen, China, 2Department of Magnetic Resonance Imaging, The First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou, China
In this work, overlapping echo acquisition together with deep learning-based reconstruction were proposed to achieve T2 mapping of abdomen in free-breathing for the first time.
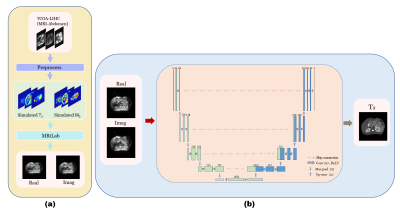
Figure 2. The flowchart of training sample generation and reconstruction of T2 mapping. (a) The single contrast abdomen MRI images from TCGA-LIHC are preprocessed first to generate T2 and M0 templates, and then the simulated MOLED acquisition is implemented on MRiLab. (b) U-Net is trained to reconstruct T2 maps from MOLED data.
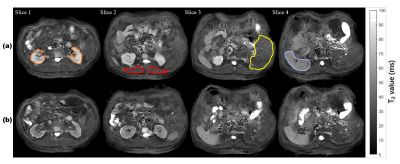
Figure 3. Reconstructed abdomen T2 maps from single-shot MOLED images. (a) T2 maps with contrast enhancement, the kidney, skeletal muscle, liver and spleen are circled in orange, red, yellow and purple, respectively. (b) T2 maps without contrast enhancement.
-
Myelin water fraction determination from relaxation times and proton density through deep learning neural network
Nikkita Khattar1, Zhaoyuan Gong1, Matthew Kiely1, Curtis Triebswetter1, Maryam H. Alsameen1, and Mustapha Bouhrara1
1Laboratory of Clinical Investigation, National Institute on Aging, Baltimore, MD, United States
An
artificial neural network model was trained and successfully used to generate
myelin water fraction maps from conventional relaxation times and proton
density maps.
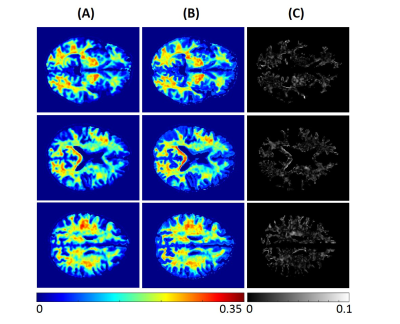
Figure 1. MWF maps from the brain imaging of a young
participant. Results are displayed for three different axial slices. (A)
represents the MWF maps calculated from BMC-mcDESPOT method (the reference
method). (B) represents MWF maps calculated using our trained neural network
(NN) model. (C) shows the absolute difference map between the reference and the
NN methods.
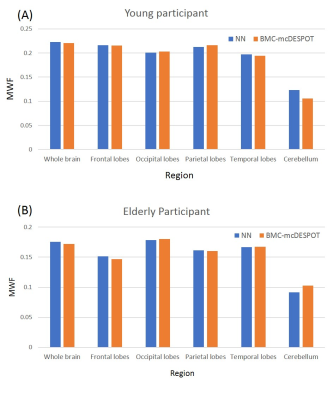
Figure 3. Mean MWF values calculated within representative
white matter brain regions using the NN (blue) and BMC-mcDESPOT (orange)
methods. Results are shown for both the young (A) and elderly (B) participants,
and indicate that NN- and BMC-mcDESPOT-derived MWF exhibit virtually similar
values for all regions evaluated.
-
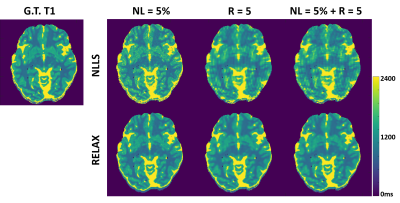
In-Vivo evaluation of high resolution T2 mapping using Bloch simulations and MP-PCA image denoising
Neta Stern1, Dvir Radunsky1, Tamar Blumenfeld-Katzir1, Yigal Chechik2,3, Chen Solomon1, and Noam Ben-Eliezer1,4,5
1Department of Biomedical Engineering, Tel Aviv University, Tel Aviv, Israel, 2Department of Orthopedics, Shamir Medical Center, Zerifin, Israel, 3Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel, 4Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 5Center for Advanced Imaging Innovation and Research (CAI2R), New-York University, Langone Medical Center, NY, United States
The Marchenko-Pastur Principle Component Analysis denoising
algorithm can be used to improve the high-resolution mapping of quantitative T2
values. The technique was validated on in
vivo brain and knee data, leading to an increase in T2 maps
precision while preserving anatomical features.
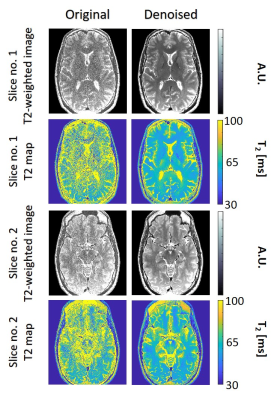
Figure
3: T2 weighted images and T2 maps for two slices acquired using the second high-resolution in vivo brain scan. Left / right columns show the pre- / post-denoising images and T2-maps (window size 15x15).
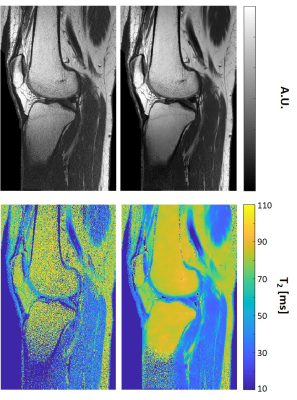
Figure
4: T2 weighted images and T2 maps of a selected slice acquired
using a high resolution in vivo knee scan (matrix size=448x280, FOV=192x120 mm2).
The left column shows the original images and T2-maps,
and the right column shows the corresponding images and T2 maps after MP-PCA image denoising
(window size 20x10).
-
Rapid learning of tissue parameter maps through random FLASH contrast synthesis
Divya Varadarajan1,2, Katie Bouman3, Bruce Fischl*1,2,4, and Adrian Dalca*1,5
1Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Department of Computing and Mathematical Sciences, California Institute of Technology, Pasadena, CA, United States, 4Massachusetts General Hospital, Boston, MA, United States, 5Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States
We propose an unsupervised
deep-learning strategy that generalizes over multiple acquisition parameters and employs the FLASH MRI model to jointly estimate T1,
T2* and PD tissue parameter maps with the goal to synthesize physically
plausible FLASH signals.
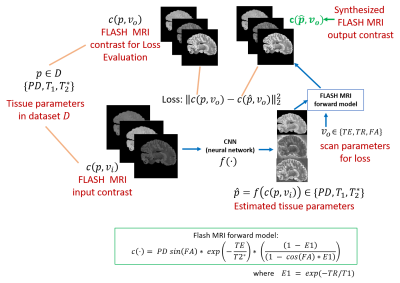
Figure 1: Proposed
framework: The proposed model to synthesize arbitrary FLASH MRI contrasts using
a CNN and a FLASH forward model from three input image contrasts. As a
consequence of using the FLASH model, the output of the CNN can be interpreted
as estimates of the tissue parameters (T1,T2* and PD).
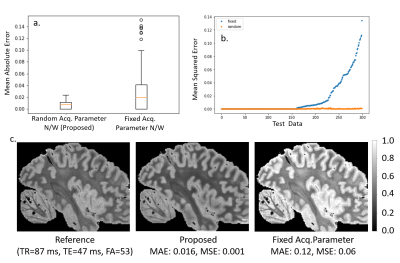
Figure
2: Contrast Synthesis: Test error in synthesis of image contrasts estimated
from three input images over 100 test datasets.
The boxplots in 2a and 2b, plot
the MAE and the images in 2c. show the reference, proposed estimate and
fixed acq. network estimate. The images in 2c. show a slice from the
test dataset, with the reference estimated from 3 flip 4 echo scan and
predicted contrast from both random and fixed networks.
-
Accurate quantitative parameter maps reconstruction method for tsDESPOT using Low Rank approximated Unet ADMM
Yuuzo Kamiguchi1, Sadanori Tomiha2, and Masao Yui3
1Advanced Technology Reserch Dept. Reserch and Development Center, Canon Medical Systems Corporation, Kawasaki, Japan, 2Advanced Technology Reserch Dept. Reserch and Development Center, Canon Medical Systems Corporation, Otawara, Japan, 3Reserch and Development Center, Canon Medical Systems Corporation, Otawara, Japan
From the data
acquired by DESPOT like sequence, full sampled low rank approximated images were estimated using ADMM method which optimize Unet estimation
and data consistency, then accurate quantitative maps were obtained by dense
neural network.
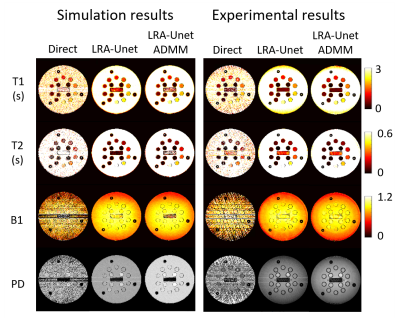
Figure 4. Representative T1, T2, B1 and PD maps of
numerical and real NIST system phantom obtained using three reconstruction
methods.
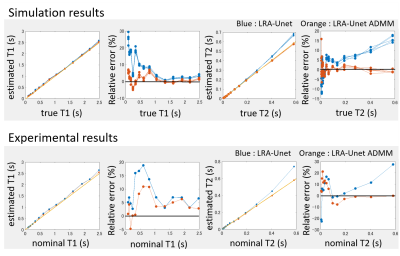
Figure 5. Mean estimated value of T1, T2, and their relative
errors in each region of numerical and real NIST system phantoms as function of
their true and nominal value each other.
-
Deep Learning Enhanced T1 Mapping and Reconstruction Framework with Spatial-temporal and Physical Constraint
Yuze Li1, Huijun Chen1, Haikun Qi2, Zhangxuan Hu3, Zhensen Chen1, Runyu Yang1, Huiyu Qiao1, Jie Sun4, Tao Wang5, Xihai Zhao1, Hua Guo1, and Huijun Chen1
1Center for Biomedical Imaging Research, Medical School, Tsinghua University, Beijing, China, 2School of Biomedical Engineering and Imaging Sciences, King’s College London, London, United Kingdom, 3GE Healthcare, Beijing, China, 4Vascular Imaging Lab and BioMolecular Imaging Center, Department of Radiology, University of Washington, Seattle, Seattle, WA, United States, 5Department of Neurology, Peking University Third Hospital, Beijing, China
A
Deep learning enhAnced T1 parameter mappIng and recoNstruction framework using
spatial-Temporal and phYsical constraint (DAINTY) was proposed. DAINTY imposed
low rank, sparsity and physical constraints to generate good quality T1
weighted images and T1 maps.
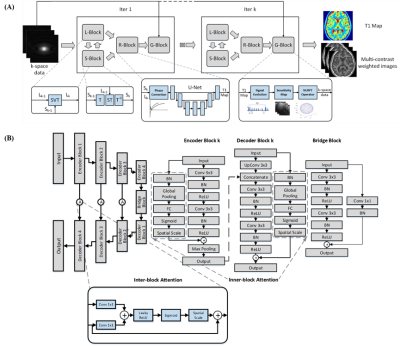
Figure 1. The illustration of DAINTY framework. (A) The under-sampled
k-space data are first processed by L-Block and S-Block to impose the low rank and
sparsity constraint. Then they are processed through R-Block to generate
refined T1 maps. The physical model G-Block is used to transform T1 map back to T1 weighted images to further improve the reconstruction quality. After k iterations the clear T1 map and high-quality T1
weighted images can be both obtained. (B) The structure of DenseAttention UNet.
The number of feature maps for encoder and decoder
block 1-4 are 256, 128, 64, and 32.
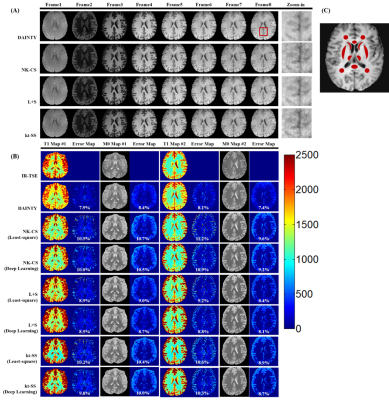
Figure 3. MR images of in-vivo human
brain. (A) T1 weighted images from GOAL-SNAP sequence using DAINTY, NK-CS,
kt-SS and L+S method; (B) Reconstructed T1 and M0 maps and error maps from
GOAL-SNAP sequence using DAINTY, NK-CS, kt-SS and L+S methods with least-square
fitting and direct deep learning mapping. T1 and M0 maps from IR-TSE are as the
reference.
-
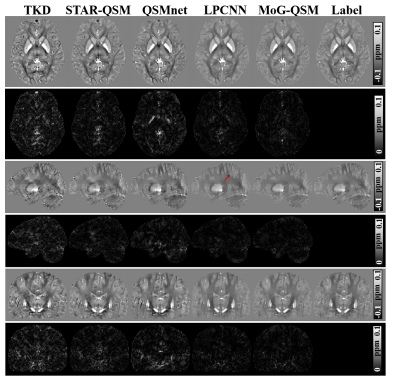
Learned Proximal Convolutional Neural Network for Susceptibility Tensor Imaging
Kuo-Wei Lai1,2, Jeremias Sulam1, Manisha Aggarwal3, Peter van Zijl2,3, and Xu Li2,3
1Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Radiology and Radiological Sciences, Johns Hopkins University, Baltimore, MD, United States
We developed a physics-informed Learned Proximal Convolutional Neural Network (LP-CNN) with specialized loss function for Susceptibility Tensor Imaging (STI) reconstruction and demonstrated its feasibility with synthetic phantoms.
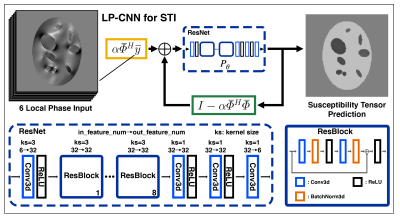
Figure 1: Network architecture of LP-CNN. In the ResNet, there are 8 stacks of residual blocks. Each residual block contains 2 convolutional layers, 2 batch normalization layers, and 2 ReLU layers with 1 skip connection for residual learning. The forward pass of LP-CNN for STI contains 3 iterations.
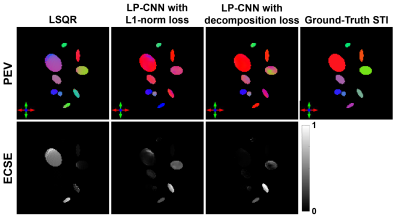
Figure 3: Principal eigenvector (PEV) maps of the reconstructed susceptibility tensors in anisotropic regions using different methods and the associated ECSE maps. The color represents the PEV direction, and the ECSE maps show the angle difference between the reconstructed PEV and the ground-truth PEV.
-
Accelerating 3D MULTIPLEX MRI Reconstruction with Deep Learning
Eric Z. Chen1, Yongquan Ye2, Xiao Chen1, Jingyuan Lyu2, Zhongqi Zhang3, Yichen Hu2, Terrence Chen1, Jian Xu2, and Shanhui Sun1
1United Imaging Intelligence, Cambridge, MA, United States, 2UIH America, Inc., Houston, TX, United States, 3United Imaging Healthcare, Shanghai, China
This is the first work applying the deep learning approach to the 3D MULTIPLEX MRI reconstruction. The proposed method shows good performance in image quality and reconstruction time.
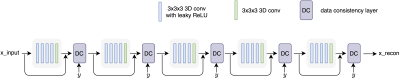
Figure 1. The network architecture for 3D MULTIPLEX data reconstruction. The model includes five convolutional blocks and each block contains five 3D convolutional layers and one data consistency layer. The feature maps are 48 for the first four 3D convolutional layers and 2 for the last 3D convolutional layer in each block. The x, y indicate image and kspace, respectively. The real and imaginary numbers of complex values are transformed into two channels and fed into the network.
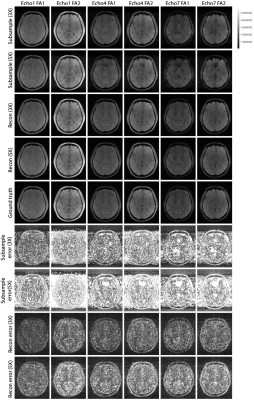
Figure 2. Examples of reconstructed MULTIPLEX echo images by the proposed deep learning method at 3X and 5X accelerations. FA1 and FA2 indicate two different flip angle configurations. Three (Echo1, Echo4 and Echo7) out of seven echo configures are showed due to space constraints. All errors are multiplied by 50 for better visualization. The same 2D axial slice from each 3D image is plotted.
-
Accelerated cardiac T1 mapping using attention-gated neural networks
Johnathan Le1,2, Jason Mendes2, Mark Ibrahim3, Brent Wilson3, Edward DiBella1,2, and Ganesh Adluru1,2
1Department of Biomedical Engineering, University of Utah, Salt Lake City, UT, United States, 2Utah Center for Advanced Imaging Research, University of Utah, Salt Lake City, UT, United States, 3Department of Cardiology, University of Utah, Salt Lake City, UT, United States
By using an attention-gated multi-layer perceptron, T1 mapping sequences can potentially be accelerated by reducing the number of T1-weighted images required to produce high-quality T1 maps.
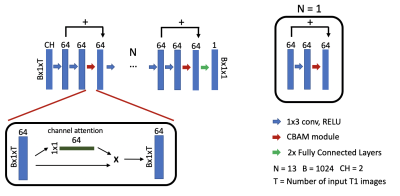
Figure 1. Illustration of the proposed attention-gated neural network for cardiac T1 mapping. Inputs to the network have dimensions of B x CH x 1 x T. CH = 2 corresponds to T1 weighted values and their inversion times. T corresponds to the number of input T1 images and inversion times for pre-contrast (T = 5) and post-contrast (T = 4) acquisitions. B = 1024 is the batch size. Scanner generated T1 maps were used as the reference.
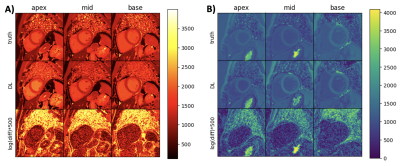
Figure 2. Network generated T1 maps in comparison to scanner generated reference T1 maps and their corresponding difference images for (A) pre-contrast T1 maps and (B) post-contrast T1 maps from two test patients. T1 maps are shown in milliseconds.
-
8X Accelerated Intervertebral Disc Compositional Evaluation with Recurrent Encoder-Decoder Deep Learning Network
Aniket Tolpadi1,2, Francesco Caliva1, Misung Han1, Valentina Pedoia1, and Sharmila Majumdar1
1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Bioengineering, University of California, Berkeley, Berkeley, CA, United States
A recurrent encoder-decoder network predicts T2 maps from undersampled T2 echos, allowing for eightfold reduction in quantitative MRI acquisition time while showing strong correlation to ground truth, low prediction error rates, fidelity to T2 values, and retainment of textures.
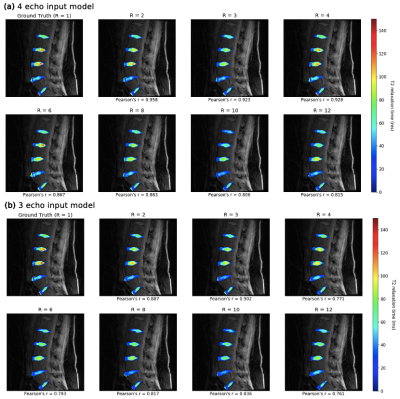
Figure 2: Performance of 3 and 4-echo pipelines in predicting fully sampled T2 maps in an example of holdout set. Pearson’s r is calculated for each map with respect to ground truth. (a) Predicted 4-echo pipeline maps showed fidelity to ground truth up to R=6, and for (b) the 3-echo pipeline, up to R=3. Up to these acceleration factors, maps reconstitute T2 values and preserve NP/AF delineation, and can thus quantitatively assess disc health and reflect early degenerative changes.
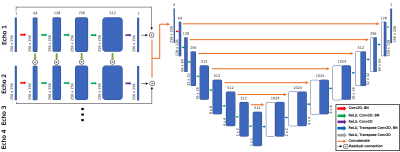
Figure 1: Recurrent encoder-decoder architecture used to predict T2 maps from spatially undersampled MAPSS T2 echos. Initial recurrent network includes connections between processing streams of each echo to exploit temporal correlations. Subsequent encoder-decoder network exploits spatial correlation and predicts final T2 map. The network could be configured for any number of input T2 echos, but the number of filters throughout the encoder-decoder network portion are presented as for 4-echo inputs.
-
Deep Learning Reconstruction of MR Fingerprinting for simultaneous T1, T2* mapping and generation of WM, GM and WM lesion probability maps
Ingo Hermann1,2,3, Alena-Kathrin Golla1,3, Eloy Martinez-Heras4, Ralf Schmidt1, Elisabeth Solana4, Sara Llufriu4, Achim Gass5, Lothar Schad1, Sebastian Weingärtner2, and Frank Zöllner1,3
1Computer Assisted Clinical Medicine, Medical Faculty Mannheim, University Heidelberg, Mannheim, Germany, 2Magnetic Resonance Systems Lab, Department of Imaging Physics, Delft University of Technology, Delft, Netherlands, 3Mannheim Institute for Intelligent Systems in Medicine, Medical Faculty Mannheim, University Heidelberg, Mannheim, Germany, 4Center of Neuroimmunology, Laboratory of Advanced Imaging in Neuroimmunological Diseases, Hospital Clinic Barcelona, Universitat de Barcelona, Barcelona, Spain, 5Department of Neurology, Medical Faculty Mannheim, University Heidelberg, Mannheim, Germany
Deep learning regression network for the reconstruction of T1, T2*,
WM, GM and white matter lesion probability maps.
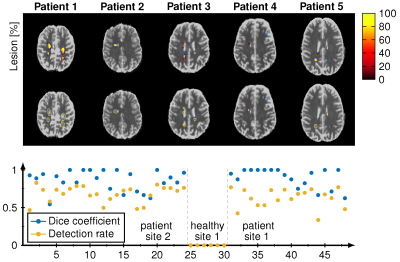
The reconstructed lesions probability maps are overlayed on the magnitude date in color encoding for all five different patients from the test set. Manual annotation is depicted in blue. Below the probability map is binarized and depicted in yellow in addition. The dice coefficient and white matter lesion detection rate is depicted for every patient and healthy subject for both sites. The average lesions detection rate is 0.83 and dice coefficient is 0.67 for all patients.
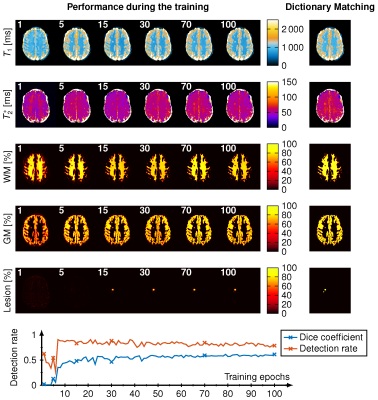
Visualization of the reconstruction during the training. The reconstructed T1, T2*, WM-, GM and Lesion-probability maps are depicted for 1, 5, 15, 30, 70 and 100 training epochs (white number) and the dictionary matching reference maps are shown on the right side. On the bottom the Dice coefficient (blue) and the lesion detection rate (orange) is depicted over the 50 training epochs.
-
The sensitivity of classical and deep image similarity metrics to MR acquisition parameters
Veronica Ravano1,2,3, Gian Franco Piredda1,2,3, Tom Hilbert1,2,3, Bénédicte Maréchal1,2,3, Reto Meuli2, Jean-Philippe Thiran2,3, Tobias Kober1,2,3, and Jonas Richiardi2
1Advanced Clinical Imaging Technology, Siemens Healthineers, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Perceptual loss is
correlated with L1 distance and outperforms other metrics in detecting changes
in acquisition parameters. Segmentation loss is poorly correlated with other
metrics, suggesting that maximizing these similarity metrics is not sufficient to
harmonize data.
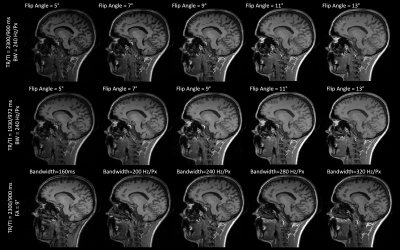
Figure 1. Contrasts obtained from
fourteen different MPRAGE protocols in one example subject. Five equally spaced
flip angles were investigated (between 5° and 13°) for two different
combinations of repetition and inversion times (TR/TI = 2300/900 ms and
1930/972 ms). Five equally spaced read-out bandwidths were also
investigated (between 160 and 320 Hz/Px) for TR/TI=2300/900 ms.
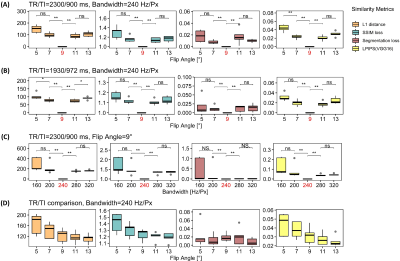
Figure 2. Variation of
similarity losses in four experimental scenarios shown in Table 2. SSIM loss is
defined as the inverse of SSIM. Segmentation loss is defined as the relative
absolute error in the thalamus volume estimation. LPIPS(VGG16) represents a learned
similarity metric based on a perceptual loss. Highlighted x-axis ticks
indicate the corresponding reference image. *: p < 0.05, **: p < 0.01,
***: p < 0.001